midiPipe, base [,files=filelist] [,mask=mask] [,smooth=smooth] [,gsmooth=gsmooth] [,/fast]
This IDL procedure actually just checks your input a little, then calls two shell scripts:
dispVis (or dispVisT if you specify /fast)
dispPhot
These scripts call a whole set of C-routines listed below.
The only required input parameter is a character string, base, which gives a prefix that will be attached to all output files produced by the pipe. This will typically also include a directory. So a typical pipe routine would be called:
inputFiles = midiGui(dir=rawdatadir)
tag = '/cool2/reduceddata/HD12345/firstTry'
midipipe, tag, file=inputFiles
If you don't specify the files input parameter, midiGui will be called from inside the pipe, but this is usually inconvenient.
mask specifies a mask file as described above. If you don't specify it, the system looks up some default masks, e.g. for PRISM data in HIGH_SENS mode, this is:
$drsRoot/software/minrts/minrts/config/minrtsMask_FIELD_PRISM_HIGH_SENS_SLIT.fits
which is of course totally transparent. Note that this is not the correct mask for data taken before December 2003!.
For old prism data, the correct mask must be typed in:
$drsRoot/software/EWS/idl/ews/midi/utilities/prismwidemask.fits
I will describe smooth and gsmooth later.
tag+'.compressed.fits'
which is in the same format as regular data (and can be accessed with oirgetdata() except that the DATA1, DATA2.. arrays are only 1 pixel high.
The actual shell call is:
oir1dCompressData "file1 file2 file3" maskfilename tag.compressed.fits
The output of this step is tag.fringes.fits,
which is also a simple imagedata structure, with now only a single DATA1 data array.
oirFormFringes tag.compressed.fits tag.fringes.fits smooth
It is sometimes desired to read out the tracking OPD in human units (e.g. microns), after combining all the possible delay lines. This you can do with
opd = oirgetopd(inputfile)
This works with almost all the data files during the reduction, since the OPD is copied from one to the next, but it is faster with data that is already compressed.
oirRotateInsOpd tag.fringes.fits tag.insopd.fits
The output is again an imagedata structure, but the actual data are now complex, but represented by two float numbers. To convert the IDL structure to complex values, and transpose the array so that time is in the X-direction and channel in the Y-direction, which is usually nicer for display, use pseudoComplex
rotdata = oirgetdata(tag+'.insopd.fits')
complexdata = pseudocomplex(rotdata.data1)
tvscl,abs(complexdata[indgen(1000),*])
In the fast version, which is somewhat less tested than the older version, the smooth in done in time space and the value of gsmooth is the standard deviation of a gaussian in seconds; the default is 0.1 sec.
The best choice of gsmooth depends on the source and the weather. Strong sources in rapidly varying OPD weather should use a small value of gsmooth, but probably not smaller than 2 frames. Alternatively weak sources in good weather should use a larger value. For weak sources in bad weather, you're out of luck.
oirGroupDelay tag.insopd.fits tag.groupdelay.fits -s gsmooth
The fast version is:
oirMTGroupDelay tag.insopd.fits tag.groupdelay.fits -s gsmooth
The result, tag.groupdelay.fits contains two interesting tables. The main data table is a pseudocomplex data set, with delay function (complex) as a function of delay (the actual value of delay as a function of pixel number is too difficult to explain here).
There is also a DELAY table in this file, which you can access with:
delay = oirgetdelay(tag+'.groupdelay.fits')
This table has two columns: the time in the usual units, and the estimated Group Delay in meters, but if you use oirgetdelay you get the answer in microns. It is very useful to plot Group Delay against tracking OPD, because if they don't follow each other, the on-line tracking mechanism failed, and your data are probably worthless.
So you can look at the data that the delay finding algorithm tried to use by
delayfunction = oirgetdata(tag+'.groupdelay.fits')
df = pseudocomplex(delayfunction.data1)
tvscl,abs(df[1000+indgen(1000),*])
tvscl,rebin(float(df[1000+indgen(5000),*]), 1000, 512)
Note: that the data returned by oirgetdata has not been smoothed in the time direction, so the S/N is lower and the time resolution higher than in the search algorithm. In particularly you will also see the image peak zooming up and down in delay as the piezos scan. If you average these in time they will get much weaker. You can try:
dfsmooth = csmooth2(df,s) {\rm s is the $\sigma$ of a gaussian smoothing kernel}
tvscl,rebin(float(dfsmooth[1000+indgen(5000),*]), 1000, 512)
It is a very good idea to look at these delay functions. If you do not see a smooth, well behaved peak, your data are probably worthless!
/oirRotateGroupDelay tag.fringes.fits tag.groupdelay.fits tag.ungroupdelay.fits
/oirRotateTGroupDelay tag.fringes.fits tag.groupdelay.fits tag.ungroupdelay.fits
There are no optional inputs, and the outputs are again pseudocomplex.
A large difference between the tracking OPD and the Group Delay OPD indicates that the on-line MIDI system had not (yet) found the fringe. The maximum allowable difference depends on the spectral resolution of MIDI being used: spectral resolution divided by the OPD difference, measured in compatable units, should be less than one radian. In practice this means that ΔOPD should be less than 100 μ for the PRISM and about 500μ for the GRISM. In particular, there are 50 or so frames at the beginning of each tracking run where we deliberately set the OPD way off, and these should be rejected from the estimation.
Secondly, if there is a big OPD jump from one frame to the next, caused e.g. by the UT auto focusing mechanism, then the data near that jump is probably garbage, because the OPD was varying substantially during single integrations. Thus the call goes:
oirAutoFlag tag.ungroupdelay.fits tag.delayfile.fits tag.flag.fits deltaOpd jumpOpd sideDrop
where deltaOpd is the allowable maximum $\Delta$OPD (microns), jumpOpd is the maximum allowable jump (microns) and sideDrop is the number of points to drop on each side of a jump. The current defaults are:
deltaOpd = 150 microns ({\rm PRISM})
deltaOpd = 800 microns ({\rm GRISM})
jumpOpd = 10 microns ({\rm from one frame to the next})
sideDrop = 1
The output file tag+'.flag.fits' contains a FLAG table as described in the original OIR FITS document, and is essentially a list of times that contain data flagged as nogood, and the reason why.
oirAverageVis tag.ungroupdelay.fits tag.flag.fits tag.corr.fits
The output: tag+'.corr.fits' is in OI_VISIBILITY format, at discussed above.
oirChopPhotometry "Afile1 Afile2" "Bfile1 Bfile2" maskFile tag.photometry.fits
For chopped photometric files with either the A or B shutter closed, the sky frames and target frames are averaged separately, and the average sky is subtracted from the average target frames. Then the data are compressed in the Y-direction in a variety of fashions. This yields a 1-dimension imagingdata file with 6-rows, each for 1 variation, plus 6-rows of error estimates, not particularly reliable, in a file called tag+'.photometry.fits':
oirRedCal tag
where the tag should be enough to point to the two files needed. The output is automatically named tag.redcal.fits.
At this point midiPipe is finished and will have put two plots on the IDL screen:
midiPipe,caltag,calfiles...
which produces caltag.redcal.fits and all the other files. We then type:
midiCalibrate, tag, calTag [,calFlux10=cf10] [,diam=d] [/print]
[, photoTag=pt] [,/nopoint]
This calls the C-program oirCalibrateVis which divides a lot of things by a lot of other things and produces at least three output files:
tag.calvis.fits (a VISIBILITY files with calibrated visibilities})
tag.calphot.fits (a DATA files with photometric fluxes in Janskys })
The first file contains the calibrated visibility amplitude and phase, which are just the uncalibrated values (as a complex phasor) divided by the values for the calibrator. The second file contains two rows:
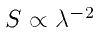 .
.
If you know that your calibrator is somewhat resolved, you can specify its diameter as diam=diameter, in millarcsec, and a uniform disk model will be used during the calibration.
The program produces a large postscript files tag+'calPlots.ps' which plots just about all the calibrated and uncalibrated quantities against wavelength, with somewhat unreliable error estimates. Look at these carefully!
The /print option causes the program to produces about 12 ASCII files named with tag----.dat, which contain the same information as the plots in case you don't want to deal with the binary FITS files for further processing of your data.
The /noPoint option is perhaps well named, and its primary use is where the target is resolved by the single telescopes. In this case a different scaling is used to estimate the correlated fluxes.