[Top] [Prev] [Next] [Bottom]
15.1 NICMOS Data Files
STScI automatically processes and calibrates all NICMOS data and archives the data files resulting from pipeline processing in FITS format. If you have retrieved NICMOS files from the Archive (see Chapter 1), you will notice that their names look like this:
n3w2a1wqm_cal.fits
The first part of the file name (n3w2a1wqm) is the rootname, identifying the dataset to which the file belongs, the second (cal) is the suffix, identifying the type of data the file contains, and the third (fits) indicates that this is a FITS format file. Chapter 2 shows how to access the data contained in NICMOS FITS files; Appendix B explains how to decipher the rootnames of these files and explains why some of them are grouped into data associations. This section describes the different types of files that constitute a NICMOS dataset.
15.1.1 File Name Suffixes
Each file in a NICMOS dataset has a three-character suffix that uniquely identifies the file contents. Table 15.1 lists the file name suffixes for NICMOS and the corresponding file contents. The files that contain final calibrated data (which you will most likely use for analysis) are highlighted. This table lists all of the files that the pipeline can produce. For some observing strategies not all of the processing steps are performed and only a subset of these files will be produced by the pipeline. A brief explanation of the contents and origin of each file is given below.
- Raw Science File (_raw):
This FITS file contains the raw image data received from the spacecraft. One file per exposure is created (a MULTIACCUM exposure is considered a single exposure irrespective of the number of samples specified).
- Support File (_spt):
This FITS file contains supporting information abou the observation, the spacecraft telemetry and engineering data from the instrument that was recorded at the time of the observation.
- Association Table (_asn):
This file is a STSDAS table that contains the list of datasets making up an association.
- Calibrated Science File (_cal):
This FITS file contains the calibrated science data for an individual dataset, and is produced by the pipeline calibration task calnica (see Chapter 16). The input to calnica are the _raw images. For a MULTIACCUM exposure, this file contains a single science image formed by combining the data from all samples.
- Intermediate Multiaccum Science File (_ima):
This FITS file is also produced by the pipeline task calnica and contains the calibrated science data for all samples of a MULTIACCUM dataset before the process of combining the individual readouts into a single image has occured. This file is only produced for MULTIACCUM observations.
- Mosaic Files (_mos):
These FITS files contain the composite target and, for chopped pattern sequences, background region images constructed by the pipeline task calnicb for an associated set of observations (see Chapter 16). The inputs to calnicb are the calibrated _cal images from calnica. Target images are co-added and background-subtracted. The value of the last character of the rootname is 0 for targets, and 1 to 8 for background images. These files are only produced for an associated set of observations.
- Post-calibration Association Table (_asc):
This table is produced by the pipeline calibration task calnicb, and is the same as the association table _asn, with the addition of new columns which report the offsets between different images of the mosaic or chop pattern as calculated by the pipeline, and the background levels computed for each image. This file is only produced for an associated set of observations.
- Trailer File:
This file contains a log of the pipeline calibration processing that was performed on individual datasets and mosaic products.
- Processing Data Quality File (_pdq):
This ASCII file provides quality information on the observation, mostly on pointing and guide star lock. Possible problems encountered, e.g., a loss of guide star lock or a guide star acquisition failure, are reported here.
15.1.2 Science Data Files
The *_raw.fits, *_cal.fits, *_ima.fits, and *_mos.fits files are all defined as science data files, as they contain the images of interest for scientific analysis.
File Contents and Organization
The data for an individual NICMOS science image consist of five arrays, each stored as a separate image extension in the FITS file. The five data arrays represent:
 In a MULTIACCUM exposure, the order of the imsets in the file is such that the
result of the longest integration time (the last readout performed on-board) occurs
first in the file (first imset), the one readout before the last is the seond imset, and
so on; the zeroth readout is the last imset (i.e., the order is in the opposite sense
from which they are obtained).
In a MULTIACCUM exposure, the order of the imsets in the file is such that the
result of the longest integration time (the last readout performed on-board) occurs
first in the file (first imset), the one readout before the last is the seond imset, and
so on; the zeroth readout is the last imset (i.e., the order is in the opposite sense
from which they are obtained).
Although the five science, error, data quality, samples, and integration time arrays associated with each imset are stored in a single file, they are kept separate as five individual FITS image extensions within the file. The order of the images in the FITS files is listed in Table 15.2 and shown graphically in Figure 15.1 and Figure 15.2. The examples given in Table 15.2 and Figure 15.2 refer to a MULTIACCUM image (multiple imsets), while Figure 15.1 refers to ACCUM, BRIGHTOBJ and RAMP images (one imset, namely extensions 1 through 5). Each extension comes with its own header, and each FITS file contains, in addition, a primary header (primary header-data unit or HDU).
The only contents of the primary header are header keywords. There is no image data in the primary header. The keywords in the primary header are termed global keywords because they apply to the data in all of the file extensions. The organization and location of header keywords is explained in detail later in the chapter.
NICMOS Science Data File Contents
Figure 15.1: Data Format for ACCUM, RAMP, BRIGHTOBJ and ACQ Modes
Figure 15.2: MULTIACCUM Mode Data Format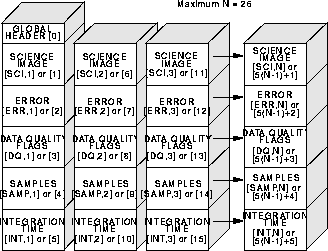
The following sections explain the contents and origin of each of the five image arrays in each imset in more detail.
Science Image
This image contains the data from the detector readout. In ACCUM, RAMP, and BRIGHTOBJ modes the image received from the instrument is the result of subtracting the initial from the final readouts of the exposure. In MULTIACCUM mode the images received are the raw (unsubtracted) data corresponding to each detector readout. In this case the subtraction of the zeroth read is done in the ground calibration pipeline task calnica (see Chapter 16). In raw datasets the science array is an integer (16-bit) image in units of DNs (counts). In calibrated datasets it is a floating-point image in units of DNs per second (count rates).
Error Image
The error image is a floating-point array containing the statistical uncertainty associated with each corresponding science image pixel. For all observing modes except RAMP, this image is computed in the ground calibration pipeline task calnica as a combination of detector read noise and Poisson noise in the accumulated science image counts (see Chapter 16) and is expressed in terms of 1 uncertainties. In RAMP mode the instrument computes the variance for each pixel value from the intermediate readouts and returns the variance image to the ground where it is stored in the raw science data file (*_raw.fits). In this case the ground calibration pipeline simply converts the variance values in the raw data file to standard deviations.
uncertainties. In RAMP mode the instrument computes the variance for each pixel value from the intermediate readouts and returns the variance image to the ground where it is stored in the raw science data file (*_raw.fits). In this case the ground calibration pipeline simply converts the variance values in the raw data file to standard deviations.
Data Quality Image
This integer (unsigned 16-bit) array contains bit-encoded data quality flags indicating various status and problem conditions associated with corresponding pixels in the science image. Because the flag values are bit-encoded, a total of 16 simultaneous conditions can be associated with each pixel. Table 15.3 lists the flag values and their meanings.
Number of Samples Image
The SAMP image is an integer (16-bit) array containing the total number of data samples that were used to compute the corresponding pixel values in the science image. For RAMP mode observations this information is computed by the instrument and sent to the ground to be recorded in the raw data file (*_raw.fits). For ACCUM and BRIGHTOBJ modes, the number of samples contributing to each pixel is set to a value of 1 in the raw data file. For MULTIACCUM mode the sample values in the raw and intermediate data files are set to the number of readouts that contributed to the corresponding science image. Because the number of samples in the raw images for MULTIACCUM, ACCUM and BRIGHTOBJ modes have one number for all pixels of an imset, the array is usually not created (to save on data volume), and the value of the sample is stored in the header keyword PIXVALUE in the SAMP image extension (see Table 15.6 below).
In MULTIACCUM calibrated data files (*_cal.fits) the SAMP array contains the total number of valid samples used to compute the final science image pixel value, by combining the data from all the readouts and rejecting cosmic ray hits and saturated pixels. In this case the sample array may have different values at different pixel locations (less or equal to the total number of samples in the MULTIACCUM sequence), depending on how many valid samples there are at each location.
In the mosaic images (*_mos.fits), the data in the SAMP array indicate the number of samples that were used from overlapping images to compute the final science image pixel value.
Integration Time Image
The TIME image is a floating-point array containing the effective integration time associated with each corresponding science image pixel value. These data are always computed in the ground calibration pipeline for recording in the raw data file. For ACCUM and BRIGHTOBJ mode observations each pixel has the same time value. For MULTIACCUM observations each pixel for a given readout has the same time value in the raw and intermediate data. Therefore, the same data-volume-saving technique is used in these cases: the array is not created and the value of the time is stored in the header keyword PIXVALUE in the TIME image extension (see Table 15.6 below). For RAMP mode observations the integration time for each pixel is computed from the exposure time per sample and the total number of samples contributing to each pixel; a TIME array is thus returned to the ground.
In MULTIACCUM calibrated data files (*_cal.fits) the TIME array contains the combined exposure time of all the readouts that were used to compute the final science image pixel value, after rejection of cosmic ray and saturated pixels from the intermediate data. As in the case of the SAMP array, the TIME array can have different values at different pixel locations, depending on how many valid samples compose the final science image in each pixel.
In mosaic images (*_mos.fits), the TIME array values indicate the total effective exposure time for all the data that were used to compute the final science image pixel values.
15.1.3 Auxiliary Data Files
The *_spt.fits, *_trl.fits, *_pdq.fits, the *_asn.fits, and the *_asc.fits files are termed auxiliary data files. They contain supporting information on the observation, such as spacecraft telemetry and engineering data, assessment of the quality of the observation, calibration information, and information on the associations present in the observations.
Association Tables
The association tables, *_asn.fits and *_asc.fits, are STSDAS tables which are created when a particular observation generates an association of datasets (see "Associations" on page B-4). In particular, the *_asn.fits table is generated by OPUS, and contains the list of datasets which make up the association (e.g., from a dither or chop pattern). The *_asn.fits tables are the inputs to the pipeline calnicb, which creates the mosaiced or background subtracted images (*_mos.fits files) from the input datasets. All the datasets must have been processed through the basic pipeline data reduction (calnica) before being processed through calnicb. In addition to the output science image(s), calnicb produces another associations table (*_asc.fits), which has the same content as the *_asn.fits table, along with additional information on the offsets used by the pipeline for reconstructing the science image. For mosaics (dither patterns), there is only one final image produced, with file name *0_mos.fits. For chop patterns, in addition to the background-subtracted image of the target (*0_mos.fits), an image for each background position will be produced; the file names of these background images are *1_mos.fits, *2_mos.fits,..., *8_mos.fits (A maximum of eight independent background positions is obtainable with the NICMOS patterns, see the NICMOS Instrument Handbook for details).
Support File
The support files *_spt.fits contain information about the observation and engineering data from the instrument that was recorded at the time of the observation. A support file can have multiple extensions within the same file; in the case of a MULTIACCUM observation there will be one extension for each readout (i.e., each imset) in the science data file. Each extension in the support file holds an integer (16-bit) image.
Trailer File
The trailer files *_trl.fits contain information on the calibration steps executed by the pipelines and diagnostics issued during the calibration.
Processing Data Quality File
The processing data quality files (*_pdq.fits) contain general information summarizing the observation, a data quality assessment section, and a summary on the pointing and guide star lock. They state whether problems where encountered during the observations, and, in case they were, describe the nature of the problem. There is one *_pdq.fits file produced for each dataset, and, in case of associations, one *_pdq file for each NICMOS product (i.e., each *_mos.fits file).
[Top] [Prev] [Next] [Bottom]
stevens@stsci.edu
Copyright © 1997, Association of Universities for Research in Astronomy. All rights
reserved.
Last updated: 11/13/97 16:58:50
 In a MULTIACCUM exposure, the order of the imsets in the file is such that the
result of the longest integration time (the last readout performed on-board) occurs
first in the file (first imset), the one readout before the last is the seond imset, and
so on; the zeroth readout is the last imset (i.e., the order is in the opposite sense
from which they are obtained).
In a MULTIACCUM exposure, the order of the imsets in the file is such that the
result of the longest integration time (the last readout performed on-board) occurs
first in the file (first imset), the one readout before the last is the seond imset, and
so on; the zeroth readout is the last imset (i.e., the order is in the opposite sense
from which they are obtained).

