[Top] [Prev] [Next] [Bottom]
2.2 FITS File Format
Flexible Image Transport System (FITS) is a standard format for exchanging astronomical data between institutions, independent of the hardware platform and software environment. A data file in FITS format consists of a series of Header Data Units (HDUs), each containing two components: an ASCII text header and the binary data. The header contains a series of header keywords that describe the data in a particular HDU and the data component immediately follows the header.
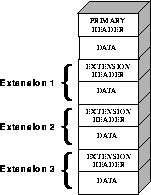
The first header in a FITS file is known as the primary header, and any number of extensions can follow the primary HDU. The data unit following the primary header must contain either an image or no data at all, but each extension can contain one of several different data types, including images, binary tables, and ASCII text tables. The value of the XTENSION keyword in the extension's header identifies the type of data the extension contains. Figure 2.1 schematically illustrates the structure of a FITS file and its extensions.
Figure 2.1: FITS File Structure
 The three-letter identifier (e.g., d0h) that follows the rootname of an HST data
file (see Appendix B for more on HST file names) has often been called an "extension" in the past. However, because of the potential for confusion with FITS
extensions, this handbook will refer to these three-letter identifiers as "suffixes."
The three-letter identifier (e.g., d0h) that follows the rootname of an HST data
file (see Appendix B for more on HST file names) has often been called an "extension" in the past. However, because of the potential for confusion with FITS
extensions, this handbook will refer to these three-letter identifiers as "suffixes."
2.2.1 Working with FITS Image Extensions
The FITS image kernel included in IRAF version 2.11 is designed to read and write the images in FITS extensions and their associated headers. Once IRAF has ingested a FITS image and its header, it treats the header-data pair like any other IRAF image. The following discussion describes how to specify the image extensions in FITS files that you would like to process with IRAF/STSDAS tasks and presumes that you are using IRAF 2.11 or higher. It covers how to:
 Retaining the .fits at the end of every FITS file name in your file specifications will ensure that IRAF both reads and writes these images in FITS format.
Retaining the .fits at the end of every FITS file name in your file specifications will ensure that IRAF both reads and writes these images in FITS format.
 If you want to work with STIS and NICMOS data, you will need to upgrade to
IRAF 2.11 or higher and STSDAS 2.0.
If you want to work with STIS and NICMOS data, you will need to upgrade to
IRAF 2.11 or higher and STSDAS 2.0.
Generating a FITS File Listing
Once you have downloaded STIS or NICMOS FITS files from the Archive, you may want an inventory of their contents. To generate a listing of a FITS file's extensions, you can use the catfits task in the tables package. The following example illustrates the first thirteen lines generated by catfits from a NICMOS MULTIACCUM FITS file containing images only.
Figure 2.2: NICMOS MULTIACCUM Listing from catfits
tt> catfits n3t501c2r_raw.fits
EXT# FITSNAME FILENAME EXTVE DIMENS BITPI OBJECT
0 n3t501c2r_raw n3t501c2r_raw.fits 16 n3t501c2r_raw.f
1 IMAGE SCI 1 256x256 16 n3t501c2r_raw.f
2 IMAGE ERR 1 -32
3 IMAGE DQ 1 16
4 IMAGE SAMP 1 16
5 IMAGE TIME 1 -32
6 IMAGE SCI 2 256x256 16
7 IMAGE ERR 2 -32
8 IMAGE DQ 2 16
9 IMAGE SAMP 2 16
10 IMAGE TIME 2 -32
11 IMAGE SCI 3 256x256 16
12 IMAGE ERR 3 -32
The first column of a catfits listing gives the extension numbers. Note that the primary HDU is labeled extension number zero. The second column lists the extension type, given by the keyword XTENSION (IMAGE = image, BINTABLE = binary table, TABLE = ASCII table). The third column lists the extension name, given by the keyword EXTNAME. In STIS and NICMOS image files, the EXTNAME values SCI, ERR, and DQ indicate science, error, and data quality images, respectively. NICMOS image files contain samples and exposure time images as well, with EXTNAME values SAMP and TIME.
Each STIS or NICMOS readout generates an image set or imset. STIS imsets comprise three images (SCI, ERR, DQ), while NICMOS imsets comprise five (SCI, ERR, DQ, SAMP, TIME). All images belonging to the same imset share the same integer value of the EXTVER keyword, given in the fourth column of a catfits listing. Several STSDAS tasks can work with entire imsets (see "Working with STIS and NICMOS Imsets" on page 3-12), but most operate on individual images. See the STIS and NICMOS Data Structures chapters for more information on the contents of imsets.
Accessing FITS Images
After you have identified which FITS image extension you wish to process, you can direct an IRAF/STSDAS task to access that extension using the following syntax:
fitsfile.fits[extension number][keyword options][image section]
Note that all the bracketed information is optional. However, the only time it is valid to provide only a file name without further specification is when the file is a simple FITS file that contains a single image in the primary HDU.
Designation of the extension number is the most basic method of access, but it is not necessary the most helpful. Referring to an extension's EXTNAME and EXTVER in the [keyword options] is often more convenient. If a number follows an EXTNAME, IRAF interprets the number as an EXTVER. For example, if extension number 6 holds the the science image belonging to the imset with EXTVER = 2, as in the catfits listing on the previous page, you can specify it in two equivalent ways:
fitsfile.fits[6]
fitsfile.fits[sci,2]
Designations giving an EXTNAME without an EXTVER refer to the first extension in the file with the specifed value of EXTNAME. For example, -fitsfile.fits[sci] refers to the first science image extension in -fitsfile.fits.
The syntax for designating image sections adheres to the IRAF standard, so in the current example the specifications
fitsfile.fits[6][100:199,100:299]
fitsfile.fits[sci,2][100:199,100:299]
both extract a 100 by 200 pixel subsection of the same science image in -fitsfile.fits.
Header Keywords and Inheritance
STIS and NICMOS data files take advantage of an IRAF image kernel convention regarding the relationship of the primary header keywords to image extensions in the same file. In particular, IRAF allows image extensions to inherit keywords from the primary header under certain circumstances. When this inheritance takes place, the primary header keywords are practically indistinguishable from the extension header keywords. This feature circumvents the large scale duplication of keywords that share the same value for all extensions. The primary header keywords effectively become global keywords for all image extensions. The FITS standard does not cover or imply keyword inheritance, and while the idea itself is simple, its consequences are often complex and sometimes surprising to users.
Generally keyword inheritance is the default, and IRAF/STSDAS applications will join the primary and extension headers and treat them as one. For example, using imheader as follows on a FITS file will print the both the primary and extension header keywords to the screen:
cl> imheader fitsfile.fits[sci,2] long+ | page
Using imcopy on such an extension will combine the primary and extension headers in the output HDU, even if the output is going to an extension of another FITS file. Once IRAF has performed the act of inheriting the primary header keywords, it will normally turn the inheritance feature off in any output file it creates unless specifically told to do otherwise.
 If you need to change the value of one of the global keywords inherited from the
primary header, you must edit the primary header itself (i.e., "extension" [0]).
If you need to change the value of one of the global keywords inherited from the
primary header, you must edit the primary header itself (i.e., "extension" [0]).
Keyword inheritance is not always desirable. For example, if you use imcopy to copy all the extensions of a FITS file to a separate output file, IRAF will write primary header keywords redundantly into each extension header. You can suppress keyword inheritance by using the NOINHERIT keyword in the file specification. For example:
im> imcopy fitsfile.fits[6][noinherit] outfile.fits
im> imcopy fitsfile.fits[sci,2,noinherit] outfile.fits
Both of the preceding commands will create an output file whose header contains only those keywords that were present in the original extension header. Note that the noinherit specification belongs bracketed with the EXTNAME and EXTVER keywords and not in a separate bracketed unit of its own.
Appending Image Extensions to FITS Files
IRAF/STSDAS tasks that produce FITS images as output can either create new FITS files or append new image extensions to existing FITS files. You may find the following examples useful if you plan to write scripts to reduce STIS or NICMOS data:
cl> imcopy fitsfile.fits[0] outfile.fits
cl> imcopy fitsfile.fits[sci,2,noinherit] \
>>> outfile.fits[append]
cl> imcopy fitsfile.fits[sci,2,noinherit] \
>>> outfile.fits[sci,3,append]
cl> imcopy fitsfile.fits[7] \
>>> outfile.fits[append,dupname]
cl> imcopy fitsfile.fits[sci,2,noinherit] \
>>> outfile.fits[sci,2,overwrite]
2.2.2 Working with FITS Table Extensions
STIS and NICMOS use FITS tables in two basic ways. Both instruments produce association tables (see "Associations" on page B-4) listing the exposures that go into constructing a given association product. In addition, STIS provides certain spectra, calibration reference files, and time-tagged data in tabular form (see Chapter 20). Here we describe:
Accessing FITS Tables
You can access data in FITS table extensions using the same tasks appropriate for any other STSDAS table, and the syntax for accessing a specific FITS table is similar to the syntax for accessing FITS images (see "Working with FITS Image Extensions" on page 2-4), with the following exceptions:
fi> catfits n3tc01010_asn.fits
EXT# FITSNAME FILENAME EXTVE ...
0 n3tc01010_asn N3TC01010_ASN.FITS ...
1 BINTABLE ASN 1 ...
Extension number 1 holds the association table, which has EXTNAME=ASN and EXTVER=1. You can use the tprint task in the STSDAS tables package to print the contents of this table, and the following commands are all equivalent:
tt> tprint n3tc01010_asn.fits
tt> tprint n3tc01010_asn.fits[1]
tt> tprint n3tc01010_asn.fits[asn,1]
STSDAS tables tasks can read both FITS TABLE and BINTABLE extensions, but they can write tabular results only as BINTABLE extensions. Tasks that write to a table in-place (i.e., tedit) can modify an existing FITS extension, and tasks that create a new table (i.e., tcopy) will create a new extension when writing to an existing FITS file. If the designated output file does not already exist, the task will create a new FITS file with the output table in the first extension. If the output file already exists, your task will append the new table to the end of the existing file; the APPEND option necessary for appending FITS image extensions is not required. As with FITS images, you can specify the EXTNAME and EXTVER of the ouput extension explicitly, if you want to assign them values different from those in the input HDU. You can also specify the OVERWRITE option if you want the output table to supplant an existing FITS extension. For example, you could type:
tt> tcopy n3tc01010_asn.fits outfile.fits[3][asn,2,overwrite]
This command would copy the table in the first extension of n3tc01010_asn.fits into the third extension of outfile.fits, while reassigning it the EXTNAME/EXTVER pair [asn,2] and overwriting the previous contents of the extension. Note that overwriting is the only time when it is valid to specify an extension, EXTNAME, and an EXTVER in the output specification.
Specifying Arrays in FITS Table Cells
A standard FITS table consists of columns and rows forming a two-dimensional grid of cells; however, each of these cells can contain a data array, effectively creating a table of higher dimensionality. Tables containing extracted STIS spectra (see "Tabular Storage of STIS Data" on page 20-5) take advantage of this feature. Each column of a STIS spectral table holds data values corresponding to a particular physical attribute, such as wavelength, net flux, or background flux. Each row contains data corresponding to one spectral order, and tables holding echelle spectra can contain many rows. Each cell of such a spectral table can contain a one-dimensional data array corresponding to the physical attribute and spectral order of the cell.
In order to analyze tabular spectral data with STSDAS tasks other than the sgraph task and the igi package, which have been appropriately modified, you will need to extract the desired arrays from the three-dimensional table. Two new IRAF tasks, named tximage and txtable, can be used to extract the table-cell arrays. Complementary tasks, named tiimage and titable, will insert arrays back into table cells. To specify the arrays which should be extracted from or inserted into the table cells, you will need to use the selectors syntax to specify the desired row and column. The general syntax for selecting a particular cell is:
intable.fits[extension number][c:column_selector][r:row_selector]
or
intable.fits[keyword options][c:column_selector][r:row_selector]
A column selector is a list of column patterns separated by commas. The column pattern is either a column name, a file name containing a list of column names, or a pattern using the IRAF pattern matching syntax. If you need a list of the column names, you can run the tlcol task (type tlcol infile.fits).
Rows are selected according to a filter. The filter is evaluated at each table row, and the row is selected if the filter is true. For example, if you specify:
infile.fits[3][c:WAVELENGTH,FLUX][r:SPORDER=(68:70)]
IRAF will extract data from the table stored in the third extension of the FITS file, infile.fits, specifically the data from the columns labelled WAVELENGTH and FLUX, and will restrict the extraction to the rows where the spectral order (SPORDER) is within the range 68-70, inclusive. Alternatively, if you specify:
infile.fits[sci,2][c:FLUX][r:row=(20:30)]
IRAF will obtain data from the table stored in the FITS file extension with an EXTNAME of SCI and EXTVER of 2. The data will come from the column FLUX and be restricted to the row numbers 20-30, inclusive. Eventually, all STSDAS and TABLES tasks will be able to use row and column selection.
[Top] [Prev] [Next] [Bottom]
stevens@stsci.edu
Copyright © 1997, Association of Universities for Research in Astronomy. All rights
reserved.
Last updated: 11/13/97 16:24:32

The three-letter identifier (e.g., d0h) that follows the rootname of an HST data file (see Appendix B for more on HST file names) has often been called an "extension" in the past. However, because of the potential for confusion with FITS extensions, this handbook will refer to these three-letter identifiers as "suffixes."
 Retaining the .fits at the end of every FITS file name in your file specifications will ensure that IRAF both reads and writes these images in FITS format.
Retaining the .fits at the end of every FITS file name in your file specifications will ensure that IRAF both reads and writes these images in FITS format.
 If you want to work with STIS and NICMOS data, you will need to upgrade to
IRAF 2.11 or higher and STSDAS 2.0.
If you want to work with STIS and NICMOS data, you will need to upgrade to
IRAF 2.11 or higher and STSDAS 2.0.
 If you need to change the value of one of the global keywords inherited from the
primary header, you must edit the primary header itself (i.e., "extension" [0]).
If you need to change the value of one of the global keywords inherited from the
primary header, you must edit the primary header itself (i.e., "extension" [0]).