 If there are discontinuities in the wavelengths, which could arise due to the
-splicing of different gratings, you should run mkmultispec in interactive mode to
verify the fits.
If there are discontinuities in the wavelengths, which could arise due to the
-splicing of different gratings, you should run mkmultispec in interactive mode to
verify the fits.
Several options for combining flux and wavelength information are available:
The mkmultispec task can put wavelength information into the flux header files in two different ways. The first involves reading the wavelength data from the .c0h file, fitting the wavelength array with a polynomial function, and then storing the derived function coefficients in the flux header file (.c1h) in multispec format. Legendre, Chebyshev, or cubic spline (spline3) fitting functions of fourth order or larger produce essentially identical results, all having rms residuals less than 10-4 Å, much smaller than the uncertainty of the original wavelength information. Because these fits are so accurate, it is usually unnecessary to run the task in interactive mode to examine them.
If there are discontinuities in the wavelengths, which could arise due to the -splicing of different gratings, you should run mkmultispec in interactive mode to verify the fits.
Because mkmultispec can fit only simple types of polynomial functions to wavelength data, this method will not work well with FOS prism data, because of the different functional form of the prism-mode dispersion solution. For prism spectra, use the header table mode of mkmultispec (see below) or create an STSDAS table using imtab.
Be aware that there is a physical limit to the number of header lines that can be used to store the wavelength array (approximately 1000 lines). This limit cannot be overridden. Under ordinary circumstances this limitation is not an issue. However, if many spectral orders have been spliced together, it may not be possible to store the actual wavelength array in the header, and a fit must be done instead.
cl> imtab y0cy0108t.c0h[8] y0cy0108t.tab wavelengthThe last word on each command line labels the three columns "wavelength", "flux", and "error".
cl> imtab y0cy0108t.c1h[8] y0cy0108t.tab flux
cl> imtab y0cy0108t.c2h[8] y0cy0108t.tab error
Constructing tables is a necessary skill if you plan to use certain tasks-such as those in the STSDAS fitting package-that do not currently recognize the multispec format WCS header information. Tabulating your spectra is also the best option if you want to join two or more spectra taken with different gratings into a single spectrum covering the complete wavelength range. Because the data are stored as individual wavelength-flux pairs, you do not need to resample, and therefore degrade, the individual spectra to a common, linear dispersion scale before joining them. Instead, you could create separate tables for the spectra from different gratings, and then combine the two tables using, for example, the tmerge task:
cl> tmerge n5548_h13.tab,n5548_h19.tab n5548.tab appendNote that you will first have to edit out any regions of overlapping wavelength from one or the other of the input tables so that the output table will be monotonically increasing (or decreasing) in wavelength.
cl> tomultispec myfile_x1d.fits new_ms.imhNote that the .imh suffix on the output file specifies that the output file is to be an OIF file. This format is similar to GEIS format, in that it consists of two files: a header file (.imh) and a binary data file (.pix). The output format for tomultispec will always be OIF.
If you want to select particular spectral orders, rather than writing all the orders to the multispec file, you will need to use the selectors syntax. To select only the spectrum stored in row nine of the input table, the previous example would change to:
cl> tomultispec "myfile_x1d.fits[r:row=9]" new_ms.imhNote that the double quote marks around the file name and row selector are necessary to avoid syntax errors. To select a range of rows, say rows nine through eleven, you would type:
cl> tomultispec "myfile_x1d.fits[r:row=(9:11)]" new_ms.imhYou can also select rows based upon values in some other column. For example, to select all rows whose spectral order lies in the range 270 to 272, type:
cl> tomultispec "myfile_x1d.fits[r:sporder=(270:272)]" \The calibrated flux is extracted by default. However, other intensity data can be specified by setting the flux_col parameter.
>>> new_ms.imh
Be careful not to restrict the search for matching rows too heavily.
Column selectors cannot be used with tomultispec.
Choose the type of fitting function for the tomultispec dispersion solution with care. Using the table option, which writes the entire wavelength array to the image header for each order, will fail if more than about three orders are selected. This restriction results from a limit to the number of keywords that can be used to store the dispersion relation.
tt> txtable "data.fits[1][c:WAVELENGTH,FLUX][r:sporder=68]" \This command would write the wavelength and flux arrays to the columns of the output table out_table. To specify multiple rows in a tabulated echelle spectrum, you would type:
>>> out_table
tt> txtable "data.fits[1][c:WAVELENGTH,FLUX][r:row=(10:12)]" \This command would generate three separate output files named echl_r0010.tab, echl_r0011.tab, and echl_r0012.tab.
>>> echl
See the online help for more details on txtable and the selectors syntax, and remember to include the double quotation marks.
The similar tximage task can be used to generate single-group GEIS files from STIS data, which can then be used as input to tasks such as resample.
tt> tximage "data.fits[1][c:WAVELENGTH][r:row=4]" wave.hhh
tt> tximage "data.fits[1][c:FLUX][r:row=4]" flux.hhh
splot
The splot task in the IRAF noao.onedspec package is a good general analysis tool that can be used to examine, smooth, fit, and perform simple arithmetic operations on spectra. Because it looks in the header for WCS wavelength information, your file must be suitably prepared. Like all IRAF tasks, splot can work on only one group at a time from a multigroup GEIS file. You can specify which GEIS group you want to operate on by using the square bracket notation, for example:
cl> splot y0cy0108t.c1h[8]If you don't specify a group in brackets, splot will assume you want the first group. In order to use splot to analyze your FOS or GHRS spectrum, you will first need to write the wavelength information from your .c0h file to the header of your .c1h files in WCS, using the mkmultispec task (see page 3-21).
The splot task is complex with many available options described in detail in the online help. Table 3.6 summarizes a few of the more useful cursor commands for quick reference. When you are using splot, a log file saves results produced by the equivalent width or de-blending functions. To specify a file name for this log file, you can set the save_file parameter by typing, for example:
cl> splot y0cy0108t.c1h[8] save_file=results.logIf you have used tomultispec to transform a STIS echelle spectrum into .imh/.pix OIF files with WCS wavelength information (see page 3-23), you can step through the spectral orders stored in image lines using the ")", "(", and "#" keys. To start with the first entry in your OIF file, type:
cl> splot new_ms.imh 1You can then switch to any order for analysis using the ")" key to increment the line number, the "(" key to decrement, and the "#" key to switch to a specified image line. Note the beam label that gives the spectral order cannot be used for navigation. See the online help for details.
3.5.4 STSDAS fitting Package
The STSDAS fitting package contains several powerful and flexible tasks, listed in Table 3.7, for fitting and analyzing spectra. The ngaussfit and nfit1d tasks, in particular, are very good for interactively fitting multiple Gaussians and nonlinear functions, respectively, to spectral data. These tasks do not currently recognize the multispec WCS method of storing wavelength information. They recognize the simple sets of dispersion keywords such as W0, WPC and CRPIX, CRVAL, and CDELT, but these forms apply only to linear coordinate systems and therefore would require resampling of your data onto a linear wavelength scale before being used. However, these tasks do accept input from STSDAS tables, in which you can store the wavelength and flux data value pairs or wavelength, flux, error value triples ("imtab" on page 3-22).
fi> ngaussfit n4449.hhh linefits.tabThis command reads spectral data from the image n4449.hhh and stores the results of the line fits in the STSDAS table linefits.tab. After you start the task, your spectrum should appear in a plot window and the task will be left in cursor input mode. You can use the standard IRAF cursor mode commands to rewindow the plot, resticting your display to the region around a particular feature or features that you want to fit. You may then want to:
 command) over which the fit will be computed so that the task will not try to fit the entire spectrum.
command) over which the fit will be computed so that the task will not try to fit the entire spectrum.
 keystroke.
keystroke.
 keystroke to redraw the screen and see the baseline that you've just defined.
keystroke to redraw the screen and see the baseline that you've just defined.
 .
.
 to compute the fit once you've marked all the features you want to fit.
to compute the fit once you've marked all the features you want to fit.
Note that when the ngaussfit task is used in this way (i.e., starting with all default values), the initial guess for the FWHM of the features will be set to a value of one. Furthermore, this coefficient and the coefficients defining the baseline are held fixed by default during the computation of the fit, unless you explicitly tell the task through cursor colon commands1 to allow these coefficients to vary. It is sometimes best to leave these coefficients fixed during an initial fit, and then to allow them to vary during a second iteration. This rule of thumb also applies to the setting of the errors parameter which controls whether or not the task will estimate error values for the derived coefficients. Because the process of error estimation is very CPU-intensive, it is most efficient to leave the error estimation turned off until you've got a good fit, and then turn the error estimation on for one last iteration.
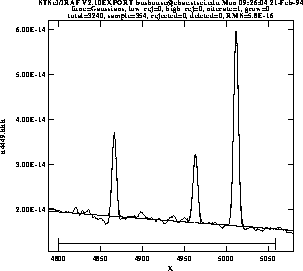
Figure 3.6 shows the results of fitting the H![]() (4861Å) and [OIII] (4959 and 5007 Å) emission features in the spectrum of NGC 4449. The resulting coefficients and error estimates (in parentheses) are shown in Figure 3.7.
(4861Å) and [OIII] (4959 and 5007 Å) emission features in the spectrum of NGC 4449. The resulting coefficients and error estimates (in parentheses) are shown in Figure 3.7.
function = Gaussians
coeff1 = 8.838438E-14 (0.) - Baseline zeropoint (fix)
coeff2 = -1.435682E-17 (0.) - Baseline slope (fix)
coeff3 = 1.854658E-14 (2.513048E-16) - Feature 1: amplitude (var)
coeff4 = 4866.511 (0.03789007) - Feature 1: center (var)
coeff5 = 5.725897 (0.0905327) - Feature 1: FWHM (var)
coeff6 = 1.516265E-14 (2.740680E-16) - Feature 2: amplitude (var)
coeff7 = 4963.262 (0.06048062) - Feature 2: center (var)
coeff8 = 6.448922 (0.116878) - Feature 2: FWHM (var)
coeff9 = 4.350271E-14 (2.903318E-16) - Feature 3: amplitude (var)
coeff10 = 5011.731 (0.01856957) - Feature 3: center (var)
coeff11 = 6.415922 (0.03769293) - Feature 3: FWHM (var)
rms = 5.837914E-16
grow = 0.
naverage = 1
low_reject = 0.
high_reject = 0.
niterate = 1
sample = 4800.132:5061.308
The input spectrum to specfit can be either an IRAF image file or an ASCII file with a simple three-column (wavelength, flux, and error) format. If the input file is an IRAF image, the wavelength scale is set using values of W0 and WPC or CRVAL1 and CDELT1. Hence, for image input, the spectral data must be on a linear wavelength scale. In order to retain data on a non-linear wavelength scale, it is necessary to provide the input spectrum in an ASCII file, so that you can explicitly specify the wavelength values associated with each data value. The online help explains a few pieces of additional information that must be included as header lines in an input text file.
2 Additional information is available in the Astronomical Data Analysis Software and Systems III, ASP Conference Series, Vol. 61, page 437, 1994.
stevens@stsci.edu Copyright © 1997, Association of Universities for Research in Astronomy. All rights reserved. Last updated: 11/13/97 16:26:00