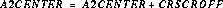[Top] [Prev] [Next] [Bottom]
21.4 Descriptions of Calibration Steps
In this section we provide a more detailed description of the algorithms applied by calstis. As always, a given step will be performed on your data if the corresponding calibration switch in the input data was set to PERFORM (see Chapter 2). The algorithmic descriptions below are described according to the major component of the calstis pipeline in which they are used, namely:
- Two-dimensional image reduction, including basic 2-D reduction, cosmic ray rejection and image co-addition.
- Processing of the contemporaneously obtained wavecal.
- Two-dimensional and one-dimensional spectral extraction, with flux and wavelength calibration.
Within each component, the individual steps are listed alphabetically, because the order in which they are performed can change for different types of data (e.g., CCD or MAMA, spectroscopic or imaging, CR-split or not).
More detailed descriptions can be found in a series of Instrument Science Reports (ISRs) that discuss the pipeline. Be aware, however, that while these reports describe the original design of the pipeline and the associated algorithms in detail, they do not always contain information concerning later modifications. Over time these reports will be updated to include a more complete description of the pipeline. In the meantime we refer you to the STIS WWW page, where a history of the important changes to the calstis pipeline code is maintained.
21.4.1 Two-Dimensional Image Reduction
ATODCORR
This applies only to CCD data. An analog to digital correction would be applied if the CCD electronic circuitry which performs the analog to digital conversion were biased toward the assignment of certain DN (data number) values. Ground test results show that this correction is not currently needed, so the ATODCORR switch will always be set to OMIT.
BIASCORR
This step is performed only for CCD data. The BIASCORR step removes any two-dimensional additive stationary pattern in the electronic zeropoint of each CCD readout after the BLEVCORR step is applied. To remove this pattern a bias reference image is subtracted. The bias reference file (BIASFILE) is a full-format superbias image created from many bias frames to assure low noise. If the science image is a subarray or is binned, a section of the bias image is extracted and binned to match the science image, prior to bias subtraction. If a CCD gain other than one is used, the bias reference file is scaled by the gain factor from the CCDTAB reference table prior to subtraction. The bias image has an associated data quality image extension: bad pixels in the bias image are flagged in the science data quality image.
BLEVCORR
This step is performed only for CCD data. The BLEVCORR step subtracts the electronic bias level for each line of the CCD image and trims the overscan regions off of the input image, leaving only the exposed portions of the image.
Because the electronic bias level can vary with time and temperature, its value is determined from the overscan region in the particular exposure being processed. A raw STIS CCD taken in full frame unbinned mode will have 20 rows of virtual parallel overscan in the AXIS2, or image y, direction, which is created by over-clocking the readout of each line past its physical extent, and 19 leading and trailing columns of serial physical overscan in the AXIS1 or image x direction, which arise from unilluminated pixels on the CCD. Thus the size of the uncalibrated and unbinned full frame CCD image is 1062 (serial) by 1044 (parallel) pixels, with 1024 x 1024 exposed science pixels.
Only the serial (physical) overscan is used for the overscan bias level determination; the virtual parallel overscan is not used. A line-by-line subtraction is performed in the following way. An initial value of the electronic bias level, or overscan, is determined for each line of the image, using only the physical serial overscan, and a function, currently a straight line, is fit to these values as a function of image line. The actual overscan value subtracted from an image line is the value of the linear fit at that image line. The initial value for each line is found by taking the median of a predetermined subset of the trailing serial overscan pixels. Currently, that region includes most of the trailing overscan region, however the first and last two pixels are skipped, as they have been shown to be subject to problems, and pixels flagged as bad in the input data quality flag are also skipped. The region used changes with binning or subarray use (see Table 21.2). The mean value of all overscan levels is computed, and the mean is written to the output SCI extension header as MEANBLEV.
In addition to subtracting the electronic bias level, the BLEVCORR step also trims the image of overscan. The sizes of the overscan regions depend on binning and whether the image is full-frame or a subimage. The locations of the overscan regions depend on which amplifier was used for readout. The number of pixels to trim off each side of the image (before accounting for readout amplifier) is given in Table 21.3. The values of NAXIS1, NAXIS2, BINAXIS1, and BINAXIS2 are obtained from image header keywords. Because the binning factor does not divide evenly into 19 and 1062, when on-chip pixel binning is used the raw image produced will contain both pure overscan pixels, overscan plus science pixels and science pixels. The calstis pipeline will only calibrate pixel binnings of 1, 2, and 4 in either AXIS1 or AXIS2.
The CRPIXi and LTVi keywords are updated in the output; these depend on the offset from removing the overscan.
CRCORR
The CRCORR step is applicable only to CCD data. This step sums the individual CR-split exposures in an associated dataset, producing a single cosmic ray rejected file (_crj.fits). The ocrreject task in the calstis pipeline is similar to the WFPC2 crrej task, except that ocrreject uses the input data quality flags to discard pixels from the input images when forming the output image, it outputs an error array for the cosmic ray rejected image using the input error arrays, and it reads the controlling input parameters (SCALENSE, INITIAL, SKY, SIGMAS, RADIUS, PFACTOR, BADINPDQ, and MASK) from the CRREJTAB reference file.
The CRCORR step does the following:
- Forms a stack of images to be combined (the CR-split or repeatobs exposures in the input file).
- Forms an initial guess image (minimum or median).
- Forms a summed CR-rejected image, using the guess image to reject high and low values in the stack, based on sigma and the radius parameter which governs whether to reject neighboring pixels to pixels identified as cosmic ray (see below).
- Performs one or more rejection cycles, using different (usually decreasing) rejection thresholds, producing a new guess image at each iteration.
- Produces a final cosmic ray rejected image, including science, data quality and error extensions, which is the weighted sum of the input images.
- Flags the data quality arrays of the individual (non-CR-rejected) input files to indicate where an outlier has been found (pixels which were rejected because of cosmic ray hits can be identified by looking for data quality bit = 14 in the _flt.fits file).
The cosmic-ray-rejected image is created by setting the value at each pixel to the sum of the values of all good pixels in the stack whose values are within plus or minus sigmas*noise of the initial guess image. Deviant (out of range) stack pixels are flagged as cosmic ray impacted by setting their stack data quality flags to 8192 in the input file.
The value of noise (in DN) is computed as: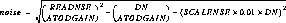
Where:
- DN = the data number of the stack pixel value.
- READNSE is the read noise in electrons, read from the primary header.
- ATODGAIN is the calibrated conversion from electrons to DN, read from the primary header.
- SCALENSE is an input parameter, read from the crrejtab -calibration -reference file.
The sigmas parameter is read from the crrejtab calibration reference file; sigmas is a string, e.g., sigmas = "4,3". The number of entries in the string dictates the number of iterations to be performed (in this example two) and the values in the string indicate the value of sigmas for each iteration. In this example, stack values that deviate from the guess image value by more than 4*noise in the first iteration are considered to be outliers and are excluded from the average on the first iteration, and an improved guess image is formed. A second iteration is then performed in which sigmas is set to 3 and good stack values disparate by more than +/- 3*noise from the guess image are excluded when determining the average. In each iteration, if RADIUS is >=1, then pixels neighboring rejected pixels are also excluded in forming the average.
SCALENSE is a string containing a multiplicative factor in the noise relation. If SCALENSE = "2.0", then the term 0.02*value is added to the noise. This term accounts for multiplicative effects, such as would be expected if this rejection were applied to Flatfield ed data. It is important to include to properly flag well-exposed regions (such as the centers of stars, where jitter from the telescope may slightly change the pointing from image to image).
The combination of the individual CRSPLIT or REPEATOBS exposures into a single cosmic-ray rejected frame is performed early in the calstis flow. The cosmic ray rejection is performed after each exposure has had its data quality file initialized and the overscan bias level subtraction (BLEVCORR) performed upon it, but prior to subtraction of a bias frame (BIASCORR), dark (DARKCORR) and flatfielding of the data (FLATCORR). The CR-rejected, bias level subtracted image is then passed through the remainder of the two-dimensional image reduction (calstis-1) to produce a flatfield ed CR-rejected image (*_crj.fits). This CR-rejected flatfield ed image is then passed through the subsequent processing steps in calstis. If EXPSCORR is set to PERFORM (see below), then the individual flatfielded but not cosmic ray rejected exposures are also produced.
DARKCORR
The DARKCORR step removes the dark signal (count rate created in the detector in the absence of photons from the sky) from the uncalibrated science image. If the science image is a subarray or was binned, the relevant section of the dark reference image must be extracted and binned to match the science image. If Doppler correction was applied on-board for the science data (i.e., if DOPPON = T), the Doppler smearing function is computed and convolved with the dark image to account for the contributions of various detector pixels to a particular image pixel. This applies only to MAMA data taken with the first order medium resolution gratings or in the echelle gratings. The Doppler convolution is done before binning the dark image. The science data quality file is updated for bad pixels in the dark reference file.
The mean of the dark values subtracted is written to the SCI extension header with the keyword MEANDARK. For CCD data, the dark image is multiplied by the exposure time and divided by the atodgain (from the CCD parameters table) before subtracting. For MAMA data, the dark image will be multiplied by the exposure time before subtracting; it will also be convolved with the Doppler smoothing function if DOPPCORR is PERFORM.
DOPPCORR
This step is performed only for spectroscopic data taken with the MAMA detectors. When MAMA data are taken in ACCUM mode in the first order medium (M) gratings or the echelle modes, the MAMA flight software corrects the location of each photon for the Doppler shift induced by the spacecraft motion, prior to updating the counter in the ACCUM mode image being produced. Therefore, during basic two-dimensional image reduction of the MAMA data, the darks and flats must be processed with the same Doppler smoothing as the science data prior to application of the reference image.
The first step is to compute an array containing the Doppler smearing function. The expression below gives the computed Doppler shift, where the time t begins with the value of the header keyword EXPSTART and is incremented in one-second intervals up to EXPSTART + EXPTIME inclusive. At each of these times, the Doppler shift in unbinned pixels is computed as: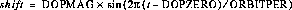
The value of shift is rounded to the nearest integer.
DQICORR
The DQICORR step takes the initial data quality file output for the science data and bit-wise ORs it with the values in the bad pixel reference file table (BPIXTAB) to initialize the science data quality file for propagation through subsequent steps in calstis. If DOPPFLAG=T, calstis will combine data quality information from neighboring pixels to accomodate Doppler smearing prior to performing the OR operation with the (unsmeared) science input data quality image. The DQICORR step also appropriately combines data quality flags in neighboring pixels if the images are binned.
EXPSCORR
If the EXPSCORR calibration switch in the header is set to PERFORM, the pipeline will also process the SCI extensions in the *_raw.fits files as individual exposures through calstis, outputting an intermediate product, *_flt.fits. This file contains the individual flatfielded CRSPLIT exposures in successive imsets of a single file. This file will not be passed through the subsequent calibration steps (e.g., spectroscopic reduction), but will be retained as an intermediate data product, to allow users to examine the effects of the pipeline cosmic-ray rejection and to re-perform the cosmic ray rejection and subsequent calibration steps as desired.
FLATCORR
The FLATCORR step corrects for pixel-to-pixel and large-scale sensitivity gradients across the detector by dividing the data by a flatfield image. The flatfield image used to correct the data is created from three flatfield reference files:
- pfltfile - This flat is a configuration (grating, central wavelength and detector) dependent pixel-to-pixel flatfield image, from which any large-scale sensitivity variations have been removed (i.e., it will have a local mean value of unity across its entirety). Such configuration dependent flats are expected to be produced infrequently, perhaps once per year.
- dfltfile - This file is a delta flat which gives the changes in the small scale flatfield response relative to the pixel to pixel flat (pfltfile). Delta flats will be taken relatively frequently (approximately monthly, though less frequently for the MAMAs); there will be a single delta flat for each detector, CCD, NUV-MAMA, and FUV-MAMA. They will be used only if needed.
- lfltfile - This flat is a subsampled image containing the large-scale sensitivity variation across the detector. It is usually grating- and central wavelength-dependent (for spectroscopic data) and aperture (filter) dependent for imaging data.
To flatfield science data, calstis creates a single flatfield image from these three files1 as described below and then divides the science image by the flat so created. The pixels of the science data quality file are updated to reflect bad pixels in the input reference files and the errors in the science data are updated to reflect the application of the flat. Blank values of pfltfile, dfltfile, or lftfile in the science data, indicate that type of flat is not to be used.
To create the single combined flatfield file, calstis first expands the large-scale sensitivity flat (lftfile) to full format. The pixel-to-pixel flat, delta flat, and expanded low-order flat are then multiplied together. For MAMA data, the product of the flatfield images will be convolved with the Doppler smoothing function if DOPPCORR = PERFORM. If a subarray or binning was used, after taking the product of all the flatfields that were specified, a subset is taken and binned if necessary to match the uncalibrated image, and the uncalibrated data are then divided by the binned subset.
GEOCORR
Geometric correction is applicable to all ACCUM mode imaging and spectroscopic data, but imaging data is not currently rectified because suitable reference files do not yet exist.
The method used is similar to 2-D rectification of spectroscopic data (see "X2DCORR" on page 21-29). For each pixel in the output rectified image, the corresponding point is found in the input distorted image, and bi-linear interpolation is used on the four nearest pixels to determine the value to assign to the output. Mapping from an output pixel back into the input images specified by two-dimensional Chebyshev polynomials stored in the format generated by the IRAF gsurfit package.
GLINCORR and LFLGCORR
These steps are performed only for the MAMA detectors. The MAMAs are photon counting detectors. At high photon (pulse) rates, the MAMA response becomes nonlinear due to three effects:
- Pore paralysis in the micro channel plates arises when charge cannot flow rapidly enough to replenish channels whose electrons have been depleted due to high local photon rates. This depletion produces a local non-linearity. The local count rate is roughly linear up to counts rates of ~200 counts/second/pixel and then turns directly over, showing an inverted V shape. Thus it is not possible to reliably correct for or flag pixels which have exceeded the local linearity limit in the pipeline (because the relation is bi-valued).
- The electronic processing circuitry has a dead-time of roughly 350 nano-seconds between pulses; thus at global count rates (across the detector) of 300,000 counts (pulses) per second, the electronic circuity counts roughly 90% of the pulses.
- The MIE electronics and flight software can process at most 300,000 pulses per second (i.e., it is matched to the expected global count rate performance of the electronic circuitry). At count rates higher than this, the MIE will still count only 300,000 pulses per second-this represents a hard cutoff beyond which no information is available to allow correction to the true count rate. In practice, at count rates approaching 270,000 counts/sec the flight software begins losing counts due to the structure of its data buffers. Further work is needed to understand this effect.
For subarrays, the hard cutoff limit of the MIE electronics and software will differ from that for full frame processing, but will still be dependent on the total global rate in addition to the rate within the subarray. The calstis pipeline currently applies the full frame correction to subarray data.
The global count rate (across the entire detector) is determined as part of the bright object protection sequence and is passed down with the exposure as a header keyword, GLOBRATE in the science header. If either GLINCORR or LFLGCORR is PERFORM, the global count rate will be checked; a correction for global non-linearity applied if GLINCORR is PERFORM, using the parameters GLOBAL_LIMIT, LOCAL_LIMIT, TAU, and EXPAND read from the mlintab reference table.
If the value of the SCI extension header keyword GLOBRATE is greater than GLOBAL_LIMIT, the keyword GLOBLIM in the SCI extension header will be set to EXCEEDED; otherwise, GLOBLIM will be set to NOT-EXCEEDED, and a correction factor will be computed and multiplied by each pixel in the science image and error array. The correction factor is computed by iteratively solving GLOBRATE = X * exp (-TAU * X) for X, where X is the true count rate. This algorithm has not yet been updated to account for the linearity effects from the flight software data buffer management.
If LFLGCORR is PERFORM, each pixel in the science image is also compared with the product of LOCAL_LIMIT and the exposure time EXPTIME. That count rate limit is then adjusted for binning by dividing by the pixel area in high-res pixels. If the science data value is larger than that product, that pixel and others within a radius of EXPAND high-res pixels are flagged as nonlinear. Because our understanding of the MAMA processing electronics is currently incomplete, accurate fluxes (global linearity) at count rates exceeding 270,000 count/sec cannot be expected from the calstis pipeline.
LORSCORR
This step is performed for MAMA data only. MAMA data are, by default, taken in high resolution mode (2048 x 2048 pixels), in which the individual microchannel plate pixels are subsampled by the anode wires. This mode produces an image with improved sampling but with appreciably worse flatfielding properties (see the STIS Instrument Handbook for more details). If LORSCORR is set to PERFORM, calstis simply adds the counts in pairs of adjacent pixels to produce images in the native format (or so-called reference format) of the MAMA detectors, with 1024 x 1024 pixels.
The binning of the uncalibrated image is determined from the LTM1 and LTM2 keywords in the SCI extension header of the raw data file. LTMi = 1 implies the reference pixel size, and LTMi = 2 means the pixels are subsampled into high-res format. In this step, if either or both axes are high-res, they will be binned down to low-res. The binning differs from binning reference files to match an uncalibrated image, in that the pixel values in this step are summed rather than averaged.
PHOTCORR
This step is applicable only for OBSTYPE=IMAGING data. For image mode, the total system throughput is read in from the phottab reference table. The photometric keywords PHOTFLAM, PHOTBW, and PHOTPLAM are computed using a synphot routine to determine the inverse sensitivity, reference magnitude, pivot wavelength, and rms bandwidth. Each quantity is written to a keyword in the primary header.
RPTCORR
This step is applicable only for MAMA data. If the number of repeat exposures is greater than one, then calstis will sum the final calibrated output file-either the flatfielded data in the case of image mode data (producing a _sfl.fits file) or the two-dimensionally extracted data (producing a _sx2.fits file) in the case of long-slit data. RPTCORR just applies a straight pixel-to-pixel addition of the science values, bit-wise ORs the data quality files and determines the error as the square root of the sum of the squares of the errors in the individual exposures.
SHADCORR
This step applies only to CCD data, but it is not currently performed. It is designed to correct for shading by the CCD shutter in very short integration time exposures. The STIS CCD shutter is specified to produce exposure non-uniformity less than or equal to 5 milliseconds for any integration time: the shortest possible STIS CCD exposure time is 100 milliseconds. Ground testing has shown that this step is not currently required.
21.4.2 WAVECAL Processing
WAVECORR
This step applies only to spectroscopic data. The purpose of wavecal processing is to determine the shift of the image on the detector along each axis owing to uncertainties in positioning by the Mode Select Mechanism (MSM) and to thermal motions. It requires one or more contemporaneous wavecal (line lamp) observations, taken without moving the MSM from the setting used for the science data.
Basic two-dimensional image reduction (basic2d) is first applied to the wavecal. For CCD data taken with the hole in the mirror (HITM) system the external shutter is ordinarily open, so the detector will have been exposed to radiation from both the science target and the line lamp. In this case, the next step is to scale the flatfield ed science image by the ratio of exposure times and subtract it from the flatfield ed wavecal. Two-dimensional rectification (x2d, see X2DCORR below) is then applied to the flatfielded (and possibly science subtracted) wavecal.
Because wavecal data are not CR-split, cosmic rays must be identified and eliminated by looking for outliers within columns, i.e., in the cross-dispersion direction. Since the data have been rectified, the image can be collapsed along columns to get a long-slit integrated spectrum or along rows to get an outline of the slit (in the cross-dispersion direction).
The shift in the dispersion direction is found by cross-correlating the observed wavecal spectrum with a template spectrum. In the cross dispersion direction, edge location is used for medium and long slits, and cross correlation is used for very short, echelle, slits. The long slits have two occulting bars, and it is the edges of these bars that are used for finding the location. Edges are found by convolving the cross-dispersion profile with the array [-1, 0, +1]. The peak in cross correlation and the edge location are obtained to subpixel level by fitting a quadratic polynomial to the three pixels nearest the extremum.
The shifts are initially measured in units of pixels of the wavecal image, but they are then scaled (depending on the binning of the wavecal) to the reference pixel size (unbinned CCD or low-res MAMA). They are subsequently written to the extension header of the 2-D rectified wavecal in the keywords SHIFTA1 (the shift in pixels along AXIS1, or dispersion direction) and SHIFTA2 (the shift in pixels along AXIS2, or spatial direction). The SHIFTA1 and SHIFTA2 keyword values are also copied from the 2-D rectified wavecal file to the flatfielded science extension header. This is the final step performed on the science data prior to 2-D rectification or 1-D extraction of the science data in the pipeline.
Either or both the wavecal file and science file can contain multiple exposures, and the image can drift across the detector over time due to such things as, thermal effects, so it is necessary to select the most appropriate wavecal exposure for each science exposure. Currently, the wavecal exposure nearest in time to a given science exposure is the one selected. Future enhancements may include interpolation of the appropriate shift.
21.4.3 Spectral Extraction
All of the following steps are applicable only to spectroscopic mode data.
BACKCORR
This step applies to one-dimensional spectral extraction only. If the calibration switch BACKCORR is PERFORM the background is calculated and subtracted from the extracted spectrum. The background is extracted above and below the spectrum, and a function is fit to the variation of the background along the spatial axis (AXIS2). The fitting function is restricted to a zeroth or first order polynomial. The polynomial order, BACKORD, is read from the XTRACTAB reference able and written to a header keyword of the same name in the output spectrum table. Average background values (in counts/sec/pixel) are calculated from each background bin, and account for fractional pixel contributions. In the case of BACKORD=0, a simple average of the two background bins is computed. For BACKORD=1, the background value at the center of each pixel that contributes to the extracted spectrum is derived from the linear fit to the background. The background in the spectrum extraction box is totaled and subtracted from the sum of the spectrum box. The total background at each pixel in the output spectrum is written to the output data table.
In general, the background or sky is not aligned with the detector pixels. To accommodate this misalignment, the definition of the background extraction apertures includes not only a length and offset (center-to-center) but also a linear tilt to assist in properly subtracting the background. This tilt is taken into account when calculating the average background in the background extraction boxes.
DISPCORR
Wavelengths are assigned using dispersion coefficients from the reference table disptab when the calibration switch DISPCORR is PERFORM; if DISPCORR is OMIT, no wavelengths are assigned. The disptab table contains dispersion solutions for a defined reference aperture. Offsets introduced by using apertures other than a reference aperture are removed using coefficients in the inangtab reference table. In the case of echelle observations, small shifts introduced by the tilt of the spectral features are removed using coefficients in the xtractab reference table.
For MAMA data, offsets of the projection of the spectrum onto the detector in both the spectral and spatial directions are deliberately introduced by offsetting the Mode Select Mechanism (grating wheel) tilts. This is done approximately monthly to assure a more uniform charge extraction from the microchannel plate over time. For MAMA observations, these induced offsets are removed using coefficients in the mofftab table.
The disptab table of dispersion contains coefficients for fits to the following dispersion solution:
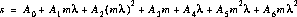
where
FLUXCORR
If FLUXCORR is PERFORM, the raw counts are corrected to 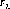 (erg cm-2 sec-1 Å-1) using the reference files PHOTTAB and APERTAB. Execution of this calibration step requires that wavelengths have been assigned. Corrections for vignetting and echelle blaze are handled within the PHOTTAB reference files. The conversion to absolute flux is calculated as:
(erg cm-2 sec-1 Å-1) using the reference files PHOTTAB and APERTAB. Execution of this calibration step requires that wavelengths have been assigned. Corrections for vignetting and echelle blaze are handled within the PHOTTAB reference files. The conversion to absolute flux is calculated as:
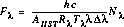
where:
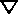 The fluxes are correct for a point source only; the flux for extended targets (as
extracted from the 2-D rectified image) requires a division by the value of the keyword DIFF2PT.
The fluxes are correct for a point source only; the flux for extended targets (as
extracted from the 2-D rectified image) requires a division by the value of the keyword DIFF2PT.
HELCORR
The correction of wavelengths to a heliocentric reference frame is controlled by calibration switches HELCORR and DISPCORR-if both switches are set to PERFORM then the correction is made. The functional form of the correction (shown below) requires the calculation of the heliocentric velocity (v) of the earth in the line of sight to the target.
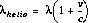
where:
SGEOCORR
This step applies only to MAMA data and is not presently performed. If SGEOCORR were PERFORM, a correction would be applied for the small scale geometric distortions in the MAMA detectors. These distortions are not adequately removed by the dispersion or spectrum or the two dimensional tracings. The corresponding reference file, SDSTFILE, contains the distortion offsets for each pixel in the MAMA image. For one-dimensional spectral extraction, all AXIS2 positions in the input image must be modified by the AXIS2 small scale distortion deltas in the small-scale distortion file. Because we do not interpolate pixels in the dispersion direction for one dimensional spectral extractions, no corrections are made to the AXIS1 positions prior to reading or extracting pixel values. Instead, the AXIS1 deltas are used to correct the assigned wavelengths.
X1DCORR
If X1DCORR is PERFORM, calstis will locate a one-dimensional spectrum to extract, and extract and flux calibrate the spectrum.
Locate the Spectrum
The nominal location of the spectrum is specified in the spectrum trace table, sptrctab and is given by (A1CENTER, A2CENTER) from this table. These coordinates are not constrained to be integers. The nominal position along the slit must be modified to include the previously updated position information found in the header. The nominal A2CENTER position of the spectrum (i.e., the position of the target along AXIS2, or the slit direction) is calculated as follows:
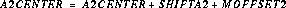
where the variables are as described in Table 21.2 sptrctab also contains the description of the distorted shape of the spectrum. The shape is stored as an array consisting of pixel offsets (in the AXIS2 direction) relative to the nominal center of the spectrum (A2CENTER). This spectrum trace is used to find, and eventually to extract, the 1-D spectrum.
The location of the spectrum is improved by searching in the vicinity of the nominal location of the spectrum by performing a cross-correlation between the distortion vector and the input spectrum image. The search extends for 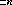 pixels around the nominal center, where n is read from the MAXSEARCH column in the xtracttab table. At each AXIS2 position in the search range (which differs from the nominal center by an integer number of pixels) a sum of the counts along the spectrum shape is formed. This sum is created by adding the value of one pixel's worth of data at each of the AXIS1 pixel positions. The pixel extracted in the AXIS2 direction is centered on the spectrum position (A2CENTER + pixel offset) and may include fractional contributions from two pixels. Quadratic refinement is used to locate the spectrum to a fraction of a pixel.
pixels around the nominal center, where n is read from the MAXSEARCH column in the xtracttab table. At each AXIS2 position in the search range (which differs from the nominal center by an integer number of pixels) a sum of the counts along the spectrum shape is formed. This sum is created by adding the value of one pixel's worth of data at each of the AXIS1 pixel positions. The pixel extracted in the AXIS2 direction is centered on the spectrum position (A2CENTER + pixel offset) and may include fractional contributions from two pixels. Quadratic refinement is used to locate the spectrum to a fraction of a pixel.
The final A2CENTER becomes:
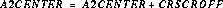
where CRSCROFF is the offset found during the cross correlation. If the cross correlation fails, the value of CRSCROFF is set to zero, a warning message is written to the output, and the A2CENTER calculated prior to the cross correlation attempt is used as the location of the spectrum. CRSCROFF is written to the output science header.
An alternate method for performing the cross correlation may be employed. In this case a 2-D template is created from the spectrum trace table. The cross correlation is carried out between the 2-D template and input image. Quadratic refinement is used as above to refine the position of the center of the spectrum to a fraction of a pixel.
Extract the 1-D Spectrum
The extraction of the spectrum is defined by a triplet of extraction boxes found in the reference table, xtractab. For each pixel in the dispersion direction, calstis sums the values in the spectrum extraction box. The extraction box is one pixel wide and has a height determined from the EXTRSIZE parameter in xtractab, centered on the spectrum. (Remember that we determined the center of the spectrum in the previous step.) The height of the extraction box may include a fractional part of one or two pixels. In the case of a fractional pixel, calstis will scale the counts in the given pixel by the fraction of the pixel extracted. Thus, each pixel in the output spectrum consists of the sum of some number (or fraction) of pixels in the input image.
The extraction of the spectrum allows for unweighted or optimal extraction. The extraction algorithm is selected based on the value of the reference table parameter XTRACALG. This flag has possible values of UNWEIGHTED and OPTIMAL. The value of XTRACALG is written to the header of the output spectrum data file. At present, calstis performs an unweighted extraction of the 1-D spectra; the optimal extraction algorithm has not yet been implemented. At the end of the 1-D extraction step, a spectrum of gross counts/second is produced.
X2DCORR
This step applies to two-dimensional spectral extraction. If X2DCORR is PERFORM, a two dimensional rectified image will be produced for spectroscopic data. The two-dimensional rectified output image (_x2d.fits or _sx2.fits) will have a linear wavelength scale and uniform sampling in the spatial direction. The dispersion direction is the first image axis (AXIS1). The size of the rectified image is made somewhat larger (the increase can be substantial for subarrays) than the input in order to allow for variations in heliocentric correction and offsets of the spectrum on the detector. The binning of the output image will be approximately the same as the input. For each pixel in the output rectified image, the corresponding point is found in the input distorted image and bi-linear interpolation is used on the four nearest pixels to determine the value to assign to the output. No correction for flux conservation is applied, as this is accounted for in the flatfield.
Mapping from an output pixel back into the input image makes use of the dispersion relation and one-dimensional trace table. The dispersion relation gives the pixel number as a function of wavelength and spectral order. The one-dimension trace is the displacement in the cross dispersion direction at each pixel in the dispersion direction. Both of these can vary along the slit, so the dispersion coefficients and the one-dimensional trace are linearly interpolated for each image line. Corrections are applied to account for image offset, binning, and subarray. The spectrum can be displaced from its nominal location on the detector for several reasons, including Mode Select Mechanism (MSM) uncertainty, deliberate offsets for distribution of charge extraction for MAMA data, and the aperture location relative to a reference aperture. These offsets are accounted for by modifying the coefficients of the dispersion relations and by adjusting the location of the one-dimensional trace. See also DISPCORR and FLUXCORR, for algorithmic details (the process of dispersion solution, spatial rectification and flux and wavelength calibration is similar for one-dimensional and two-dimensional spectral extracted data).
[Top] [Prev] [Next] [Bottom]
1
The rationale for maintaining three types of flatfield reference files rather than a single integrated reference file is described in detail in STIS ISR 95-09 "Calibration Plans for Flat Fielding STIS Data."
stevens@stsci.edu
Copyright © 1997, Association of Universities for Research in Astronomy. All rights
reserved.
Last updated: 11/13/97 17:36:49
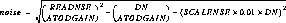
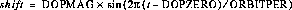
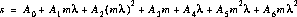
 is the wavelength in Angstroms.
is the wavelength in Angstroms.
 is the detector AXIS1 position.
is the detector AXIS1 position.
 is the spectral order.
is the spectral order.
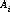 are the dispersion coefficients.
are the dispersion coefficients.
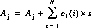
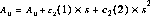
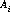 dispersion coefficients
dispersion coefficients incidence angle coefficients
incidence angle coefficients aperture offsets in the axis 1 direction calculated as difference of relative aperture centers (arcsec)
aperture offsets in the axis 1 direction calculated as difference of relative aperture centers (arcsec)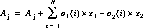
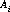 dispersion coefficients
dispersion coefficients MAMA offset coefficients
MAMA offset coefficients MAMA offset (MOFFSET1) (pixels)
MAMA offset (MOFFSET1) (pixels) MAMA offset (MOFFSET2) (pixels)
MAMA offset (MOFFSET2) (pixels)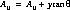
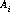 dispersion coefficients
dispersion coefficients axis 2 offset from nominal A2CENTER during spectrum locate process (pixels)
axis 2 offset from nominal A2CENTER during spectrum locate process (pixels) spectrum tilt angel
spectrum tilt angel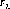 (erg cm-2 sec-1 Å-1) using the reference files PHOTTAB and APERTAB. Execution of this calibration step requires that wavelengths have been assigned. Corrections for vignetting and echelle blaze are handled within the PHOTTAB reference files. The conversion to absolute flux is calculated as:
(erg cm-2 sec-1 Å-1) using the reference files PHOTTAB and APERTAB. Execution of this calibration step requires that wavelengths have been assigned. Corrections for vignetting and echelle blaze are handled within the PHOTTAB reference files. The conversion to absolute flux is calculated as: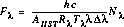
 is the calibrated flux at a particular wavelength. This quantity is also multiplied by the ATODGAIN if the data were obtained with the CCD.
is the calibrated flux at a particular wavelength. This quantity is also multiplied by the ATODGAIN if the data were obtained with the CCD.
 is Planck's constant.
is Planck's constant.
 is the speed of light.
is the speed of light.
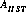 is the area of the unobstructed HST primary mirror (45238.93416 cm2).
is the area of the unobstructed HST primary mirror (45238.93416 cm2).
 is the throughput of the STIS instrument configuration at a particular wavelength when a clear full aperture is in place.
is the throughput of the STIS instrument configuration at a particular wavelength when a clear full aperture is in place.
 is a particular wavelength.
is a particular wavelength.
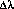 is the dispersion (Å/pixel) at a particular wavelength.
is the dispersion (Å/pixel) at a particular wavelength.
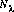 is the net count rate at a particular wavelength.
is the net count rate at a particular wavelength.
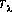 is the aperture throughput at a particular wavelength.
is the aperture throughput at a particular wavelength.
 The fluxes are correct for a point source only; the flux for extended targets (as
extracted from the 2-D rectified image) requires a division by the value of the keyword DIFF2PT.
The fluxes are correct for a point source only; the flux for extended targets (as
extracted from the 2-D rectified image) requires a division by the value of the keyword DIFF2PT.
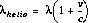
 is the heliocentric wavelength.
is the heliocentric wavelength.
 is a particular wavelength.
is a particular wavelength.
 is the component of the velocity of the earth in the direction of the target.
is the component of the velocity of the earth in the direction of the target.
 is the speed of light.
is the speed of light.
 , is written to the trailer file.
, is written to the trailer file.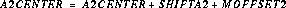
 pixels around the nominal center, where n is read from the MAXSEARCH column in the xtracttab table. At each AXIS2 position in the search range (which differs from the nominal center by an integer number of pixels) a sum of the counts along the spectrum shape is formed. This sum is created by adding the value of one pixel's worth of data at each of the AXIS1 pixel positions. The pixel extracted in the AXIS2 direction is centered on the spectrum position (A2CENTER + pixel offset) and may include fractional contributions from two pixels. Quadratic refinement is used to locate the spectrum to a fraction of a pixel.
pixels around the nominal center, where n is read from the MAXSEARCH column in the xtracttab table. At each AXIS2 position in the search range (which differs from the nominal center by an integer number of pixels) a sum of the counts along the spectrum shape is formed. This sum is created by adding the value of one pixel's worth of data at each of the AXIS1 pixel positions. The pixel extracted in the AXIS2 direction is centered on the spectrum position (A2CENTER + pixel offset) and may include fractional contributions from two pixels. Quadratic refinement is used to locate the spectrum to a fraction of a pixel.