[Top] [Prev] [Next] [Bottom]
21.5 Recalibration of STIS Data
Sometimes the default pipeline calibration, performed shortly after the data were obtained from the telescope, is not the best possible calibration for your science program. There are a number of reasons why it may be desirable to recalibrate your data. The most likely reasons include:
- More appropriate reference files have become available since the pipeline calibration was performed. CCD darks, biases, and hot pixel tables are examples of reference files that are updated frequently, but they require some time to be installed in the pipeline. Likewise we expect to be delivering updated sensitivity files during Cycle 7, updated flatfields, updated aperture throughputs, and so on. In short, over the next year or so, we expect regular improvements in reference files as we carry out the initial on-orbit calibration of STIS.
- Contemporaneous CCD flatfields were obtained with the science data for G750L or G750M NIR observations to remove fringing.
- Some steps need to be repeated with different input parameters. For example, you may wish to re-perform the cosmic ray rejection or the 1-D spectral extraction after adjusting the input parameters. The best target and background extraction regions for extracting 1-D spectra can depend on the science goals of the program. In addition, no extraction is currently performed for first-order spectral modes, so GOs with first-order spectra of standard stars will commonly also wish to perform one-dimensional extraction.
- The calibration software has been enhanced; here again we expect frequent updates over the course of the next year (see Chapter 25).
The STIS calibration pipeline was designed to accommodate the need for full or partial recalibration. As mentioned at the beginning of this chapter, calstis is re-entrant, so that certain calibration steps can be performed outside of the pipeline, and others can be executed multiple times, depending upon the science goals.
Generally, the calibration switches in the header control the operations that calstis performs on the data. There are three basic ways to select which operations are performed during calibration:
- Edit the calibration switches and run the calstis task.
- Use one or more of the pipeline subset tasks described below, managing the calibration through task parameters.
- Run the calstis sub-tasks at the host level (i.e., outside of IRAF) using the command-line switches and flags to control the processing.
This section describes the first two methods. In the end, the calibration switches in the headers of the calibrated data files will reflect the operations performed on the calibrated data and the reference files used.
21.5.1 Mechanics of Full Recalibration
You have chosen to fully recalibrate your STIS data. There is a certain amount of set-up required for calstis to run properly. The operations mentioned in the checklist below will be described in detail in the following subsections:
- Set up a directory with the required reference files.
- Set the environment variable oref to point to your reference file directory. Note: you must do this before starting an IRAF session!
- In an IRAF session, update the input data file headers using chcalpar.
- Run calstis or a subset of its constituent tasks.
Retrieve Reference Files
To recalibrate your data, you will need to retrieve the reference files used by the different calibration steps to be performed. The names of the reference files to be used during calibration must be specified in the primary header of the input files, under the section "CALIBRATION REFERENCE FILES." Note that the data headers will already be populated with the names of the reference files used during pipeline calibration at STScI.
Chapter 1 describes how to obtain the best available reference files from the HST Data Archive using StarView. For each dataset in the Archive, StarView will list both the reference files used in the initial calibration and the ones currently recommended. This list also indicates in the "Level of Change" column how much the reference files used differ from the recommended ones.
If better calibration reference files have become available since the original pipeline calibration, they can be retrieved from the HST Data Archive as explained in Chapter 1. These files might contain updated information about the instrument signatures, such as an updated hot pixel list or a bad pixel table, or an improved background bias level in a bias frame. Note that "new" does not necessarily mean that the data need to be recalibrated. Use the "Level of Change" information provided by StarView to help determine if recalibration is necessary.
The STIS reference files are all in FITS format, and can be in either IMAGE or BINTABLE extensions. The names of these files along with their corresponding primary header keywords, extensions, and format (image or table), are listed in Chapter 2. The (somewhat obscure) rootname of a reference file is based on the time that the file was delivered to the Calibration Data Base System (CDBS).
Edit the Calibration Header Keywords
To edit file headers in preparation for recalibration, use the STSDAS task chcalpar. The chcalpar task takes a single input parameter-the name(s) of the raw data files to be edited. When you start chcalpar, the task automatically determines that the data are from STIS, determines the detector used and whether the observing mode was SPECTROSCOPIC or IMAGING, and opens one of four STIS-specific parameter sets (pset) that will load the current values of all the calibration-related keywords. To edit the calibration keyword values:
- Start the chcalpar task, specifying a list of images in which you want to change calibration keyword values. If you specify more than one image, (using wildcards, for example) the task will read the initial keyword values from the first image in the list. For example, you could change keywords for all STIS raw science images in the current directory (with initial values from the first image), using the command:
ct> chcalpar o*_raw.fits
- After starting chcalpar, you will be placed in eparam-the IRAF parameter editor; from there you will be able to edit the set of calibration keywords. Change the values of any calibration switches, reference files or tables to the values you wish to use for recalibrating your data.
- Exit the editor when you are done making changes by typing ":q" two times. The task will ask if you wish to accept the current settings. If you type "y", the settings will be saved and you will return to the IRAF cl prompt. If you type "n", you will be placed back in the parameter editor to redefine the settings. If you type "a", the task will abort and any changes will be discarded.
The parameter editor screen for STIS MAMA spectroscopy is illustrated in Figure 21.11. The characters "oref$" preceding the names of the reference files specify a logical directory for the location of the reference files. The method for setting a corresponding environment variable is given in the next subsection.
Figure 21.11: Editing Calibration Keywords with chcalpar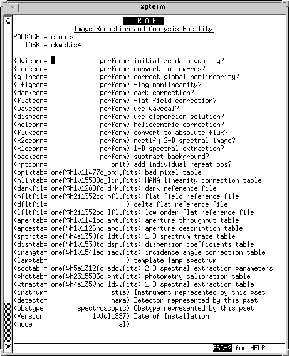
It is also possible to use hedit to update the input file keywords. The example below illustrates how to turn on the bias correction switch and update the name of the bias image reference file for all STIS raw images in the current directory that begin with the characters "o3y."
cl> hedit o3y*_raw.fits[0] biascorr PERFORM up+
cl> hedit o3y*_raw.fits[0] biasfile "oref$new_bias.fits" up+
 It is dangerous to change keyword values with hedit if the keywords reside in the
FITS primary header unit, as is the case with all calibration keywords. The correct
way to use this task on inherited keywords is to edit the primary header explicitly
by appending "[0]" to the FITS file name.
It is dangerous to change keyword values with hedit if the keywords reside in the
FITS primary header unit, as is the case with all calibration keywords. The correct
way to use this task on inherited keywords is to edit the primary header explicitly
by appending "[0]" to the FITS file name.
For each task (except calstis) it is not necessary to specify which calibration steps to perform in the primary header keywords of the input files. The execution of each step can be specified in the input parameters task of each stand-alone. The only exception is calstis where the switches in the primary header control the calibration steps to be performed.
The reference file names for all the stand-alone tasks and calstis have to be specified in their corresponding header keywords. See Table 20.5 for the list of reference files that correspond to each executable.
Running calstis in IRAF
Before running calstis, you will need to define an environment variable to indicate the location of the directory containing the needed calibration reference files. The names of the calibration files are preceded with the logical path name "oref$" in the STIS science headers. Ordinarily you would define this directory in an IRAF session to be, for example., "/data/vega3/stis/cal_ref" using the set command:
cl> set oref "/data/vega3/cal_ref/" # Won't work!
Note the trailing slash (/). However, calstis and all of its modules are actually foreign tasks and as such do not access IRAF environment variables. Therefore, before invoking the cl, you will need to define an environment variable from the host command line (see below) that is appropriate to your host machine. For Unix systems, the appropriate command for the example above is:
% setenv oref /data/vega3/cal_ref/
Then start IRAF.
 When running calstis or any of its modules, you must define environment variables (such as oref$) before starting the cl. It is not possible to define them
within IRAF using the set command, nor is it possible to define them with an
escape to the host level, such as: !setenv oref /data/vega3/cal_ref/
When running calstis or any of its modules, you must define environment variables (such as oref$) before starting the cl. It is not possible to define them
within IRAF using the set command, nor is it possible to define them with an
escape to the host level, such as: !setenv oref /data/vega3/cal_ref/
21.5.2 Rerunning Subsets of the Calibration Pipeline
Selected portions of the pipeline can be executed with special tasks in the STSDAS stis package. The tasks that can be simply used in this fashion are listed in Table 21.5 below. See also Table for the association between basic2d, ocrreject, wavecal, x1d, and x2d and the components of the calstis pipeline. When you run these tasks individually, many of the calibration parameters usually read from the reference file can be entered either as command line arguments or via epar.
The inttag task for TIMETAG data will accumulate selected events from the raw event table, writing the results as one or more image sets (imsets) in a single, output FITS file. You can optionally specify an explicit starting time, time interval, and number of intervals over which to integrate, and the collection of imsets will be written to the output file, simulating a REPEATOBS ACCUM observation. Breaking the data into multiple, short exposures can be useful not only for variables but also to improve the flatfielding when the Doppler shift is significant. Once the images have been created, it is straightforward to process them with calstis and to analyze the output image or spectra, as appropriate.
The screen messages that appear when running any calstis module are equivalent to the trailer file contents delivered with the data.
Combining Images to One File
Each calibration task (including calstis itself) takes only one science file as input, though it is sometimes useful to perform only part of calstis, such as cosmic ray rejection, on a set of images as if they were part of a repeated series. The prescription is to copy the relevant input files to a single FITS file with multiple imsets, per the format described in Figure 20.6 and Figure 20.7, and then run the relevant calibration task on the combined file. (Such a procedure is, in fact, useful for constructing bias and dark reference files for the CCD.) The easiest way to do this, while preserving the correct file format, is to use the mstools.msjoin task. For example, to combine two FITS files while removing cosmic rays:
cl> msjoin file1.fits,file2.fits big_file.fits
cl> ocrreject big_file.fits combined.fits
[Top] [Prev] [Next] [Bottom]
stevens@stsci.edu
Copyright © 1997, Association of Universities for Research in Astronomy. All rights
reserved.
Last updated: 11/13/97 17:36:49


It is dangerous to change keyword values with hedit if the keywords reside in the FITS primary header unit, as is the case with all calibration keywords. The correct way to use this task on inherited keywords is to edit the primary header explicitly by appending "[0]" to the FITS file name.
When running calstis or any of its modules, you must define environment variables (such as oref$) before starting the cl. It is not possible to define them within IRAF using the set command, nor is it possible to define them with an escape to the host level, such as: !setenv oref /data/vega3/cal_ref/