The FORS skyline alignment verification tool
The FORS skyline alignment tool can be run either within Reflex or stand-alone. The purpose of the tool is to view and modify the results of the skyline alignment done by the recipes fors_align_sky or fors_align_sky_lss, and fors_resample. In the classification tags below the MXU acronym can be read also as MOS or LSS.
Inputs of the tool
Inputs of fors_align_sky(_lss)
- SPATIAL_MAP_MXU (not in case of LSS(like) data)
- CURV_COEFF_MXU (not in case of LSS(like) data)
- DISP_COEFF_MXU
- RECTIFIED_ALL_SCI_MXU, or SCIENCE_UNFLAT_MXU in case of LSS(like) data
- SLIT_LOCATION_MXU
- MASTER_SKYLINECAT
- GRISM_TABLE
Outputs of fors_align_sky(_lss)
- SKY_SHIFTS_SLIT_SCI_MXU, or SKY_SHIFTS_LONG_SCI_MXU in case of LSS(like) data
- WAVELENGTH_MAP_SCI_MXU
- DISP_COEFF_SCI_MXU
Outputs of fors_resample
- MAPPED_ALL_SCI_MXU
Outputs of the tool
- the outputs of fors_align_sky(_lss) and fors_resample (see above) refilled according to changed slit(s)
- a fits table indicating slit(s) from where the user removed line(s)
Python script
The Python FORS skyline alignment tool consists of two files: FORSalignSky.py is a launcher script and reflex.py contains Reflex usage specific module. Both scripts are located under reflex/reflex-current/scripts/python.
Stand-alone usage
Run tool with default inputs in directory./data. You can also give the individual locations of the input files as parameter (see 'Example usage' below).
python FORSalignSky.py -d
Running fors skyline verification tool
Using default values:
{'specStep': '1', 'grismTable': './data/FORS2_GRS_300I_21_OG590_32.fits', 'mappedAll': './data/mapped_all_sci_mxu.fits',
'LineCat': './data/skylines_sampo.fits', 'invDispSol': './data/disp_coeff_mxu.fits', 'rectifiedLamp':
'./data/rectified_all_sci_mxu.fits', 'curvCoeffs': './data/curv_coeff_mxu.fits', 'residuals':
'./data/sky_shifts_slit_sci_mxu.fits', 'slitLocation': './data/slit_location_mxu.fits', 'spatialMap':
'./data/spatial_map_mxu.fits', 'wavelengthMap': './data/wavelength_map_sci_mxu.fits', 'upgradInvDispSol':
'./data/disp_coeff_sci_mxu.fits'}
{'newDispCoeffs': '/home/tester/reflex_tmp_forsSpecTool-8408/disp_coeff_mxu_new.fits', 'newResiduals':
'/home/tester/reflex_tmp_forsSpecTool-8408/sky_shifts_slit_sci_mxu_new.fits', 'newMappedAll':
'/home/tester/reflex_tmp_forsSpecTool-8408/mapped_all_sci_mxu_new.fits', 'newWavelenMap':
'/home/tester/reflex_tmp_forsSpecTool-8408/wavelength_map_sci_mxu_new.fits', 'newLineCatalog':
'/home/tester/reflex_tmp_forsSpecTool-8408/skylines_sampo_ModifiedSlits.fits'}
Other parameters
-
-helpshows input parameters -
-qmakes a Reflex query
python FORSalignSky.py -help
Available options: ['-help', '-q', '-d'] Inputs: ['specStep', 'grismTable', 'mappedAll', 'LineCat', 'invDispSol', 'rectifiedLamp', 'curvCoeffs', 'residuals', 'slitLocation', 'spatialMap', 'wavelengthMap', 'upgradInvDispSol'] Outputs: ['newDispCoeffs', 'newResiduals', 'newMappedAll', 'newWavelenMap', 'newLineCatalog'] Example usage:FORSalignSky.py wavelengthMap=parameter
The last line above means that all other input files except the wavelength map will be the defaults (i.e. run the tool with parameter -d).
Clean-up
Note that when using the tool stand-alone, the user has to take care of cleaning the results directory every now and then, e.g.:
rm -rf /tmp/reflex_tmp_forsSpecTool-*
Examples
The first example screenshot below is right after start-up. The example in the picture below is for the seventh row in the eleventh slit. In this example there are 21 slits containing 709 rows in total.
Below are explained all the buttons of the tool.
 for exiting the tool.
for exiting the tool.
 return all windows to original mode in current slit and image row.
return all windows to original mode in current slit and image row.
 for toggling between linear and logarithmic scale in spectrum profile window (middle right side).
for toggling between linear and logarithmic scale in spectrum profile window (middle right side).
 for moving/zooming the three right side windows in horizontal direction.
for moving/zooming the three right side windows in horizontal direction.
See below note for matplotlib pan/zoom mode.
 for moving to preceding and subsequent image row.
for moving to preceding and subsequent image row.
 for toggling between displaying slit number or slit ID in the slit buttons, current mode is painted gray.
for toggling between displaying slit number or slit ID in the slit buttons, current mode is painted gray.
- slit buttons are displayed in two columns, clicking a button will display the center image row of the chosen slit, current slit is green.

 for returning to initial values.
for returning to initial values.
 for realigning the current slit with new alignment polynomial order (AlignOrder) and/or deleted/recovered lines.
for realigning the current slit with new alignment polynomial order (AlignOrder) and/or deleted/recovered lines.
 for decreasing/increasing the polynomial order for sky lines alignment, valid range is 0-2.
for decreasing/increasing the polynomial order for sky lines alignment, valid range is 0-2.
 for toggling X-axis between pixels and wavelengths in the three right side windows, the two upper windows (single slit and spectrum profile window) are toggled between RECTIFIED_ALL_SCI_MXU (X-axis in pixels) and MAPPED_ALL_SCI_MXU (X-axis in &lambda), the next available mode is printed on the button.
for toggling X-axis between pixels and wavelengths in the three right side windows, the two upper windows (single slit and spectrum profile window) are toggled between RECTIFIED_ALL_SCI_MXU (X-axis in pixels) and MAPPED_ALL_SCI_MXU (X-axis in &lambda), the next available mode is printed on the button.
 for changing the cuts of the images in the all slits and single slit windows.
for changing the cuts of the images in the all slits and single slit windows.
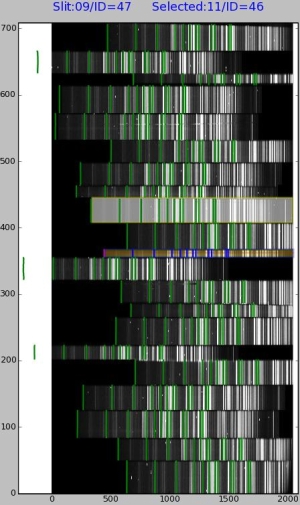
Line identification window (all slits)
Window image shows the slit locations and spectrum with identified lines which are plotted over the spectrum. The lines illuminate the grade of quality with current line catalogue and inverse dispersion solution polynomial fitting order. The goal is to find anomalies in identified line locations.
Usage

Slit number and slit identification number are shown on top of the image window. Left side shows the slit upon which the cursor is currently and right side is the selected (activated by clicking) slit.
Mouse options
When moving mouse over image the slit under mouse pointer will be highlighted and surrounded by a yellow box.
The slit can be selected by clicking the left mouse button. Selected slit will be highlighted yellow.
Current image row is shown with a red bar over the current slit.
Toolbar
The matplotlib default toolbar has options which are useful for the image zooming and panning.
 The home button will return the image as it was in launch. A known problem is that in this tool this does not always work properly (e.g. when spectrum profile window is in logarithmic scale).
The home button will return the image as it was in launch. A known problem is that in this tool this does not always work properly (e.g. when spectrum profile window is in logarithmic scale).
 Left and right arrows will work as a 'undo' and 'redo' when zooming or panning image.
Left and right arrows will work as a 'undo' and 'redo' when zooming or panning image.
 Pan with left mouse, zoom with right. NOTE: matplotlib pan/zoom mode must be applied to the single slit window, and when mouse button is released the changes done will reflect to the two windows below it keeping the X-axis scale identical in all the three right side windows. Note that the script blocks moving/zooming the single slit window in vertical direction.
Pan with left mouse, zoom with right. NOTE: matplotlib pan/zoom mode must be applied to the single slit window, and when mouse button is released the changes done will reflect to the two windows below it keeping the X-axis scale identical in all the three right side windows. Note that the script blocks moving/zooming the single slit window in vertical direction.
 Zoom area to rectangle.
Zoom area to rectangle.
Spectra of one slit, spectrum profile and results of alignment
The right-hand side of the tool window has three windows.
When X-axis is in pixels, active area of slit is painted light gray in the two lower windows.

Single slit spectrum
The upper window plots the current single slit (highlighted in the left-side window).
Current image row is shown with a red bar. Image row can be chosen by clicking on single slit window, but NOT in matplotlib pan/zoom mode.
The dispersion solutions for the identified lines calculated with the wavelength calibration polynomial coefficients are shown with blue lines.
For zoom/move buttons see above where all the buttons are explained.
When X-axis is in pixels, the single slit and spectrum profile windows contain the RECTIFIED_ALL_SCI_MXU (produced by fors_extract_slits) versus X CCD position of the current image row.
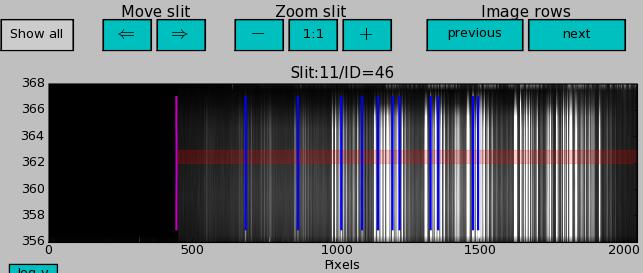
When X-axis is wavelengths, the single slit and spectrum profile windows contain the MAPPED_ALL_SCI_MXU (produced by fors_resample) versus wavelength.

Spectrum profile
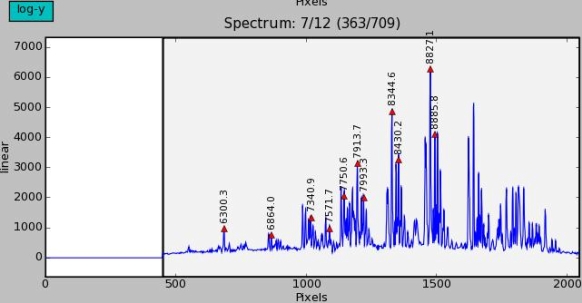
The middle window shows the spectrum profile of one image row. The current image row is printed above the window: the current slit contains 12 rows and all the slits together contain 709 rows.
The peak positions of the spectrum are marked with red triangles. The identified line wavelength is printed above the peak.
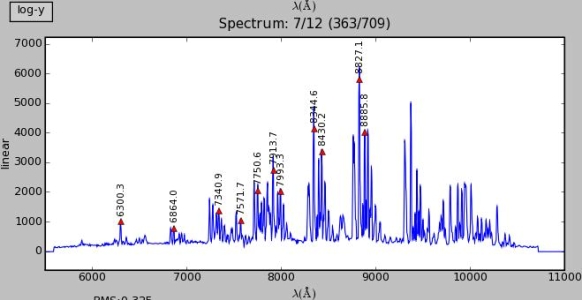
Results of alignment
The lower window plots the observed sky lines offsets of the alignment for the current slit (NOTE that unlike residuals in the wavelength calibration tool, the offsets are here per slit and not per image row).
Lines outside active area or not found by the alignment are indicated with a black triangle down in the upper edge of window.
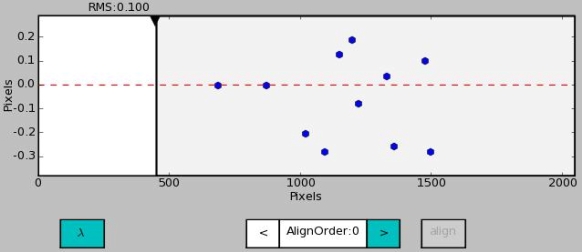
The X-axis can be toggled between pixels and wavelength (first button from the left below window).
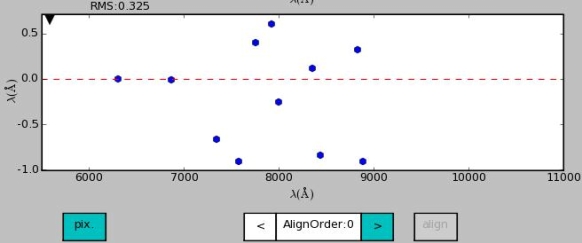
Deleting of line(s)
Click with left mouse button blue ball(s) in the lower window. The ball will turn red and activate the align button. You can cancel your intended deletion with right mouse button. The realignment is done by pressing the  button. After realignment, a red triangle down in the upper edge of the offsets window will indicate the line(s) deleted by the user.
button. After realignment, a red triangle down in the upper edge of the offsets window will indicate the line(s) deleted by the user.
In the figure below two lines with the largest offsets are marked for deletion and the align button is active.
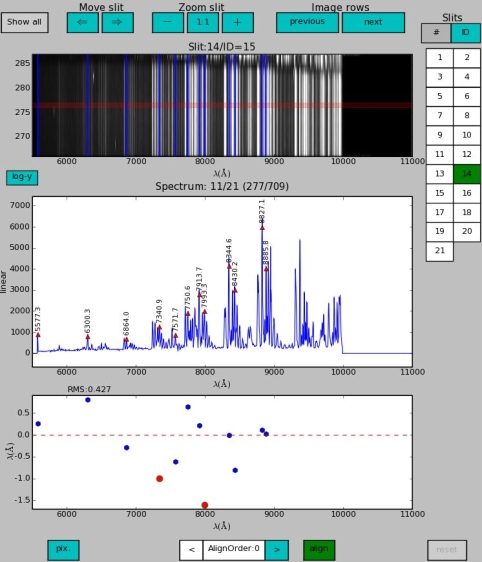
In the figure below the realignment has been done and the positions of the two deleted lines are marked with a red triangle down in the upper edge of the offsets window. The RMS improved from 0.427 Å to 0.233 Å. Consider however also this point: the more lines, the better indicator the RMS is, and the more stable the solution.

The tool produces a fits table that keeps track of the lines deleted by the user. The first column is a copy of the original line catalog. When a line is removed from a slit, a new column with the removed line(s) set to zero is added.

Recovering deleted line(s)
Deleted lines can be recovered by clicking with left mouse button the red triangle down turning it blue. You can cancel your intended recovering with right mouse button. The realignment is done by pressing the  button.
button.
In the figure below the other one of the two lines deleted in the previous step is marked for recovery and the align button is active.

Changing the polynomial order of the sky lines alignment
Click either arrow beside the AlignOrder box to decrease/increase the polynomial order. This will activate (turn green) the align button. The realignment is done by pressing the  button after which the alignment polynomial order of the current slit is stored to the new value. If you move to another image row before pressing the align button, the polynomial order will be restored to the original value and the align button deactivated.
button after which the alignment polynomial order of the current slit is stored to the new value. If you move to another image row before pressing the align button, the polynomial order will be restored to the original value and the align button deactivated.
In the figure below we first of all see that the slit button number 14 is yellow because this slit was changed in the previous step. Secondly the reset button is active (green) since we have made changes. Now we increase the alignment polynomial order of the current (slit button number 8 is green) slit to 1 and realign it. The tool runs the recipe fors_align_sky with parameter --skyalign set to AlignOrder.
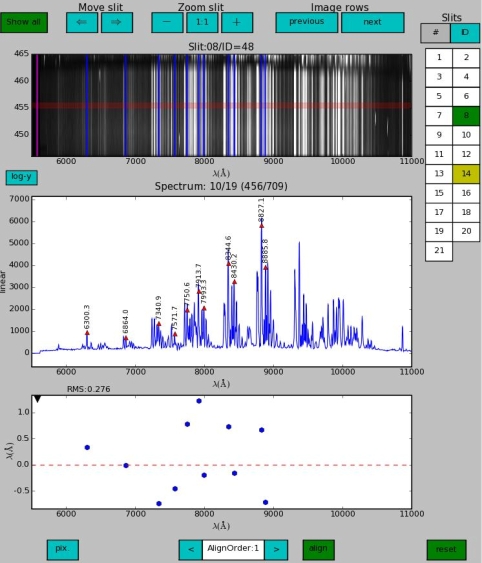
In the figure below we see that the previous step did not certainly improve the solution and moreover RMS worsened from 0.276 Å to 0.58 Å. This is no surprise since instead of determining a median offset from all observed skylines (--skyalign=0), --skyalign=1 tries to fit a slope, which is rarely useful, but sometimes sky lines offsets display a significant dependency on the wavelength.
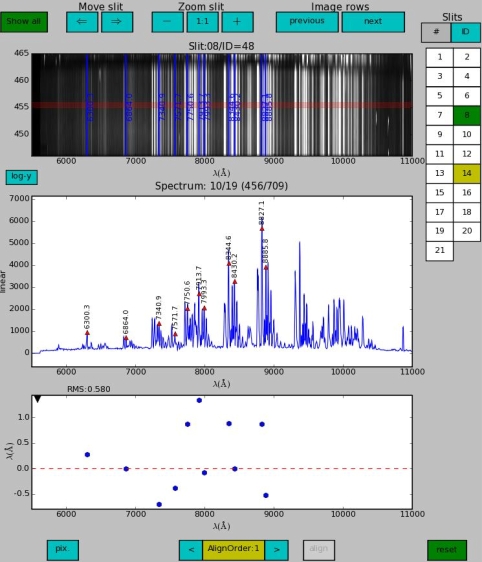
Notes
The realignment is in both cases (deleting/recovering lines and changing polynomial order) done for one slit at a time. A realigned slit is indicated by painting yellow the AlignOrder box and the slit button. After the first realignment the reset button is activated (turned green).
The combined number of blue/red balls and black/red/blue triangles down in the alignment results window equals always the number of lines in the original line catalog.
In the alignment results window red/blue balls and black/red/blue triangles down are per slit (that is they are the same for all image rows in a slit). Note the difference with the wavelength calibration tool.
